- Department of Pathobiological Sciences, Louisiana State University, Baton Rouge, LA, United States
Abstract
Bunyamwera virus (BUNV) (Bunyamwera orthobunyavirus) has been found in Sub-Saharan Africa and demonstrated recently as cocirculating with Rift Valley Fever Virus (RVFV). Little is known regarding the breadth of transmission modalities of Bunyamwera. Given its co-occurence with RVFV, we hypothesized the transmission system of BUNV shared similarities to the RVFV system including transmission by Ae. aegypti mosquitoes and environmentally mediated transmission through fomites and environmental contamination. We exposed Ae. aegypti mosquitoes to BUNV and evaluated their ability to transmit both vertically and horizontally. Further, we investigated the potential for a novel transmission modality via environmental contamination. We found that the LSU colony of Ae. aegypti was not competent for the virus for either horizontal or vertical transmission; but, 20% of larva exposed to virus via contaminated aquatic habitat were positive. However, transstadial clearance of the virus was absolute. Finally, under simulated temperature conditions that matched peak transmission in Rwanda, we found that BUNV was stable in both whole blood and serum for up to 28 days at higher total volume in tubes at moderate quantities (103–5 genome copies/mL). In addition, infectiousness of these samples was demonstrated in 80% of the replicates. At lower volume samples (in plates), infectiousness was retained out to 6–8 days with a maximum infectious titer of 104 PFU/mL. Thus, the potential for contamination of the environment and/or transmission via contaminated fomites exists. Our findings have implications for biosafety and infection control, especially in the context of food animal production.
Impact statement
In many areas where infection control and/or the use of personal protective equipment is not common, there is a risk of transmission via fomites when pathogens are environmentally stable. Our findings demonstrate that BUNV, though an RNA virus, is very environmentally stable and retains infectiousness under certain conditions. Our findings establish the proof-of-principle for this novel transmission modality of environmental contamination of BUNV.
Introduction
Bunyamwera virus (BUNV) (Bunyamwera orthobunyavirus) has been found in Sub-Saharan Africa and demonstrated recently as cocirculating with Rift Valley Fever Virus (RVFV) [1]. RVFV is a mosquito-borne arbovirus of One Health importance. Infected ruminates can experience hemorrhagic fever, spontaneous abortions, and potentially death, especially in young animals [2]. This has not only the obvious animal health implications but can be economically devastating in areas where cattle are a main source of food and/or income [3]. Humans infected with RVFV can also experience hemorrhagic fever, ocular complications, sever flu like symptoms, and death [4]. Originally isolated during an outbreak in the Rift Valley of Kenya in 1930 [5], RVFV has since been found in countries in across Africa and on the Arabian peninsula [6]. As seen with other mosquito-borne viruses, viral transmission of RVFV can often be tied to environmental factors, such as heavy seasonal rainfall and flooding events [7].
During an outbreak of RVFV in Rwanda, BUNV was found to be the etiological agent of abortions in cattle where clinical diagnosis would have attributed such symptoms to RVFV [1]. These two viruses are from the same order (Bunyavirales) and are both arboviruses transmitted by mosquitoes [8–13]. BUNV has historically been found in Sub-Saharan Africa. While BUNV can infect humans as well, causing mild symptoms including fever, myalgia/arthralgia, and rashes, the virus has been most often associated with ruminate infection and disease, including spontaneous abortions [1, 14–18]. These symptoms overlap with RVFV, though the transmission patterns of BUNV remain to be fully characterized, and is likely under surveilled. However, given the coincident transmission of RVFV and BUNV, it is reasonable to hypothesize that the BUNV transmission system may share similarities with that of RVFV.
The current understanding of the RVFV transmission cycle includes outbreak/epidemic and maintenance subcycles (Supplementary Figure S1) [9]. The maintenance cycle involves mosquitoes–believed to be including the genus Aedes–obtaining infected bloodmeals, subsequently developing a systemic infection, including the ovaries, which results in transovarial transmission [19–21]. Aedes spp. eggs are resistant to desiccation [22, 23], allowing for survival through the dry season and, the return of seasonal rains facilitate the hatch of floodwater and container Aedes mosquito eggs and the subsequent emergence of infected adults [24]. This mechanism of maintenance through the dry season then seeds a seasonal outbreak, which transitions into the outbreak subcycle, as large numbers of Culex spp. mosquitoes hatch after rain events [25, 26]. Additional transmission pathways exist that do not involve the vector. RVFV is a bloodborne pathogen and transmission to humans occurs with handling of infectious fluids from ill livestock, as well as processing raw meat [27–29]. Risk of infection also increased with handling of abortive material [28], which suggests that environmentally mediated infections are an important piece of the transmission puzzle. Adding to the risk of environmentally mediated transmission of RVFV, stability and persistence of infection was noted when originally isolated, as it was found that RVFV is stable in a citrine solution at room temperature for over 7 days [5], and subsequently, this phenomena was replicated at a wide variety of temperatures [30–32], including a potential infection occurring from a sample months old [33]. Further, contamination of larval habitat has been shown to infect larva and emerge as infected adults, indicating that environmentally mediated transmission is a non-negligible part of RVFV transmission risk [34].
Aedes aegypti, an important vector of a multitude of human pathogens, is one of the Aedes spp. competent for RVFV and BUNV, and several strains were found to be competent vectors of a closely related North American Orthobunyavirus species, Cache Valley virus [35–40]. Transovarial transmission of RVFV in Aedes aegypti was implicated in the 2007 outbreak in Sudan [41] and has been shown to be successful and stable for three ovarian cycles [20], as well as for a related Orthobunyavirus, LaCrosse virus. But transovarial transmission has not been rigorously studied in Ae. aegypti for BUNV [42]. Previously, it was demonstrated that BUNV retained infectiousness in cell culture for up to 30 days at 37°C [43]. This sustained persistence led to the consideration of whether BUNV might overlap in the environmentally mediated transmission subcycle of RVFV, as well. Herein, we explore the potential that the BUNV transmission cycle is complex and multi-faceted, like that of RVFV. We investigate the competence of Aedes aegypti for vertical and horizontal transmission, and examine factors associated with risk for environmentally mediated transmission under relevant temperature conditions.
Materials and methods
Daily Rwandan temperature profile
To simulate relevant temperature conditions, publicly available weather data was mined to generate a daily temperature profile for RVF transmission season in Rwanda, May through July, per the 2018 outbreak [1]. A data scraper collected temperature data in half-hour increments from May 18th through July 12th, 2018, and the temperatures from the same time point each day were averaged to make a single average day. The environmental chamber was then programed to mimic these conditions and a digital thermometer was placed within to confirm the temperature (Supplementary Figure S2). Humidity was maintained at approximately 85% during the experiments roughly matching historical humidity in Rwanda [44].
Viruses and mosquitoes
BUNV was obtained from the World Arbovirus Reference Center at UTMB had been passaged through suckling mouse cells 47 times and Vero cells 6–8 times prior to the experimentation. Stocks were titered via crystal violet plaque assay and determined to be ∼5.0 × 106 pfu/mL. Aedes aegypti mosquito (Rockefeller colony) eggs from were vacuum hatched for 45 min in ddH20 and allowed to emerge as in [45].
qRT-PCR and plaque assay
Viral RNA extraction was performed via MagMax 96 viral isolation as in [43]. qRT-PCR was performed on a Roche Lightcycler 96, using Quantbio Ultratough master mix and previously published primers and probes [43]. Viral RNA quantification was analyzed via comparison to previously crystal violet plaqued stock from above. Standard crystal violet plaque assays were performed [46]. Virus titer was averaged from countable plaques at each dilution [47].
qRT-PCR sensitivity assay for mosquito pools
Ae. aegypti reared as above were allowed to mature to adults. Adult mosquitoes were cold anesthetized and separated by sex and placed in to 900 µL BA-1 media with 2 stainless steel BBs in a 2 mL locking Eppendorf tube in pools of 10 and homogenized as previously described [45] and a known titer of virus was added to each tube of titers of 105, 103, 101, and 100 pfu/mL. Five replicates per pools sex were performed at each titer.
Mosquito competence and transstadial transmission
Adult Aedes aegypti females, 2 days post emergence (dpe) were offered an infectious blood meal of ratio 2:1 of bovine blood in Alsevers (Hemostat Labs, Dixon, CA, United States) and BUNV infectious supernatant (total infectious titer 1.7 × 106) via the Hemotek feeding apparatus (Discovery Labs, Blackburn, United Kingdom). Engorged females were separated by aspiration and cold anesthetization, then placed in individual 4-ounce containers and provided 10% sucrose solution ad libitum. Each container was supplied with egg paper that was moistened daily and collected when eggs were observed for an additional 18 days. Three females died prior to the end of the experiment and were collected and tested for virus in the legs and bodies as described below. At 18 days post infectious bloodmeal, mosquitoes were processed to determine infection status of bodies, legs, and saliva as in [48].
There is mixed evidence for the role of second bloodmeals in enhancing vector competence [48–51]. Thus, a second cohort of mosquitoes was offered an infectious bloodmeal as above at 3–4 days post emergence. A second uninfected bloodmeal was offered starting at 4 days following the first bloodmeal [50] and repeated attempts were made until all females had taken a second bloodmeal (maximum 9 days following first bloodmeal). The mosquitoes were maintained as described above until processed following egg collection at 22–24 post initial, BUNV infectious bloodmeal.
Environmental mediated transmission of BUNV to Ae. aegypti (transstadial transmission)
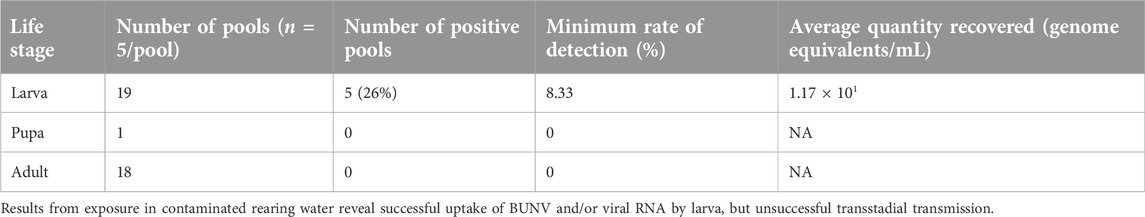
Ae. aegypti were hatched and reared as described above. Fourth instar larva were collected and rinsed in ddH20 and placed into water contaminated with BUNV. Briefly, BUNV supernatant was added to rearing water, and samples were collected at 0–5 days to establish the presence of BUNV RNA [34]. Exposed larva were allowed to mature and were collected as pupa, rinsed, and transferred to a cage for emergence. Pupa were separated based on number of days spent exposed to infected rearing water. Upon emergence, adult mosquitoes were cold anesthetized and separated by sex into pools of up to five based on exposure time. Remaining unemerged individuals were collected at 6 days post contaminated water exposure, rinsed as above, and collated into pools of up to ten for testing by qRT-PCR.
BUNV environmental stability study
To assess environmental stability of BUNV under environmentally relevant conditions, 110 𝛍L of virus was added to 4.39 mL of two different substrate types–either whole blood or serum for a final concentration of 1 × 105 pfu/mL. The virus-substrate mixture was added to individual units (either falcon tubes or 6-well plates, see Supplementary Methods) and placed into the environmental chamber under Rwanda temperature conditions. Tubes were uncapped and the six-well plate lids lifted to allow air exposure; all units were placed in secondary containment for safety and contamination avoidance. Tubes were sampled at days 0, 1, 7, 21, and 28, while plates were sampled each day until dry. Plates were sampled daily until dry, which occurred between 6 and 8 days. Collected samples were placed into a −80°C freezer until further analysis. Samples were tested starting with the latest timepoint (tubes) or with the day before the plate was dry (Dry Day −1). If a sample was found not to be infectious at a timepoint (I.e., timepoint X), the timepoint immediately preceding (i.e., timepoint X-1) was tested and this pattern continued until we found the latest time of infectivity. Ten replicates of each unit type were conducted per substrate type. Samples were assayed for stability of RNA via qRT-PCR as above and for infectivity via a growth assay on Vero cells as in [43].
Statistics
For the sensitivity assay, a Kruskal Wallace non-parametric analysis of variance was used to determine if a difference in detection ability existed between sexes of the pools. A linear regression model was used to evaluate the relationship between the initial input into the mosquito pool and the qRT-PCR output. The limit of detection (LOD) was determined using a qPCR LOD calculation R script (at 95% confidence) [52, 53]. The lowest concentration of consistently detectable RNA was determined by running multiple iterations of a standard curve assay tied to a plaque assay. The plaque assay revealed that maximum stock titer was 5.3 × 106 PFU/mL. The standard curve runs determined that the consistent point at which there was below 95% confidence detection was 5.3 × 10−1 PFU/mL. Growth was assessed via non-parametric t-test between day 1 and day 7 where a significantly higher titer at day 7 indicated growth and thus infectiousness. To evaluate stability, RNA recovered at each timepoint was compared via a Kruskal-Wallace test where day was a factor; a post hoc Dunn’s test (Bonferroni correction) test was performed where appropriate. Additionally, comparisons between RNA recovered from each substrate at the same timepoint or day until dry were done via Wilcoxon Ranked Sum tests. Significance was assessed at the α = 0.05 level. Analyses were performed using R Studio (2023.03.1 + 446) and R (4.3.0) and associated packages [54]. Minimum infection rate was determined by calculating the rate of infection as if only one individual per positive pool was infected, as in [55].
Results
Sensitivity assay
The qRT-PCR assay used herein was highly sensitive for BUNV in the context of mosquito pool testing (LOD = 6.6 copies/mL with 95% confidence). There was no significant difference in the amount of viral RNA detected in pools of males versus females (Supplementary Figure S3, p > 0.05), indicating the assay was sensitive in pools of both sexes. There was only one discrepant result where 5/5 male pools versus 4/5 female pools were positive at the lowest titer of 100 pfu/mL. Combining sexes, at titers of 101 and above, successful detection of virus was 100% (n = 10). At 100, BUNV was detected in 90% of pools (n = 10, p > 0.05).
Ae. aegypti transmission capabilities for BUNV
We found our colony of Ae. aegypti were not competent for BUNV and thus not able to horizontally or vertically transmit the virus after a single bloodmeal (n = 27). A single mosquito that died at 1 day post exposure tested positive for a midgut infection, but was not included in the analysis as this was more than likely residual RNA from the infectious bloodmeal (midgut titer 1.3 × 103 pfu/mL). Mosquitoes receiving a second bloodmeal also showed no BUNV midgut infection (n = 19).
Rearing water contaminated with BUNV remained positive for RNA for the 5 days monitored. Initial quantification of RNA copies were approximately at 104.5 genome equivalents/mL on days 0–1, while this dropped and remained consistent at 102.5–3 for days 2–5. Attempts to quantify BUNV in rearing water via plaque assay were unsuccessful. Despite detection of BUNV RNA, no adult pool tested positive for BUNV. Similarly, a single pool of two pupa that had not emerged by day 6 were also negative for virus (Table 1, see Supplementary Methods). Additionally, larva that had not pupated by day 6 were also pooled and tested. Of the 19 pools, 5 (26%) tested positive for BUNV RNA with low titer (Table 1).
Extracellular stability of BUNV RNA under ecologically relevant conditions
Under simulated Rwandan conditions, in both whole blood and serum, BUNV RNA was detectable at moderate genome copies/mL up to 28 days in tubes (Figure 1A). The difference in average titer of genome copies between whole blood and sera was statistically significant at all time points (p < 0.05). Of interest, both in whole blood and sera, BUNV RNA remained moderately high out to day 28 (6.06 × 103 and 1.89 × 105, respectively). RNA was stable over all days collected for sera. In whole blood a significant decrease in RNA compared to baseline at day 0 was observed starting on day 21, but no further significant decrease was observed between days 21 and 28 (Figure 1A, p < 0.05). Unsurprisingly, liquid volume in plates evaporated quickly relative to tubes (Supplementary Figures S4–S5) and plate wells were dry between 6 and 8 days post inoculation. The last day of sampling is referred to as Day Dry—1 (Dry-1, see Supplemental Methods). On average, 4.22 × 104 BUNV genome copies/mL was recovered on Dry-1 for whole blood, while the average recovered genome copies in sera was significantly more (p < 0.05) at 1.21 × 105 copies/mL on Dry-1 (Figure 1B). In sera the genome copies were stable as no significant differences were found between the RNA recovered on day 0 and Dry-1 (p > 0.05), while the RNA copies recovered in whole blood increased from day 0 to Dry −1 (p < 0.05). In a subset of plates (n = 5), media was added to dried out wells, collected, and subsequently tested for BUNV RNA (see Supplementary Methods). BUNV RNA was detected in the reconstituted samples, with an average of 6.87 × 104 RNA copies/mL in the reconstituted sera samples, and 6.74 × 104 RNA copies/mL in the reconstituted whole blood samples.
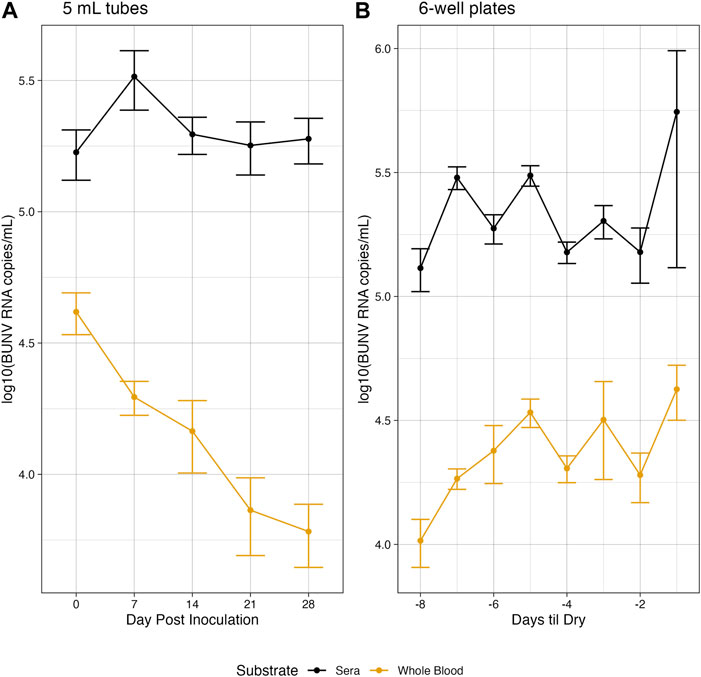
Figure 1. Environmentally stable BUNV. Genomic copies of BUNV recovered at each collection for both higher volume tubes (A) and lower volume plates (B) over time when exposed to Rwandan environmnetal temperatures.
Infectivity of stable BUNV
For whole blood in tubes, the last day of infectiousness was day 28 for 80% (8/10) of samples and day 21 for the remining 20%. In tubes containing sera, the last day of infectiousness was day 21 for 50% (5/10) of replicates while the other 50% retained infectiousness out to day 28. Cytopathic effects (CPE) were observed in all infectious samples. Attempts to plaque sera and whole blood at these later time points were unsuccessful due to the consistency of the samples (see Supplementary Methods, Supplementary Figure S4). In 6-well plates, the range of days where infectiousness was retained was 5–7 days post inoculation but was dependent on the remaining volume in the well (Supplementary Figure S3; Supplementary Table S1). Relative to the first day of no volume, 50% (5/10) of the whole blood replicates retained infectiousness at Dry-1, and 100% at Dry-2. For sera however, all replicates retained infectiousness at Dry −1. We were able to plaque plate samples to further investigate the infectious titer of environmentally stable BUNV on the last day of infectiousness. Overall, whole blood had a significantly lower average infectious titer (1.04 × 103 PFU/mL) compared to sera (2.45 × 104 pfu/mL) (Figure 2, p < 0.05). No CPE was observed in vitro for any of the reconstituted dried samples, indicating a lack of infectivity despite moderate quantitative recovery of ∼104 RNA copies/mL (see Supplementary Table S2).
Figure 2. Infectious titer of BUNV in plates. The titer of infectious BUNV recovered from plates on the last day of observed infectiousness was significantly higher for sera than for whole blood (p < 0.05) relative to the day the plate was dry (Dry Day).
Discussion
BUNV was previously found to co-circulate with RVFV, and as such BUNV has been hypothesized to share aspects of the RVFV transmission cycle [1, 8]. However, the details surrounding the modalities of BUNV transmission remains understudied. RVFV has the potential to be transmitted both vertically and horizontally by Ae. aegypti [20, 40]. Further, other Orthobunyaviruses, such Lacrosse virus, Cache Valley virus (a strain of BUNV), and others have been shown to be transmitted by Ae. aegypti [35, 36, 42, 56]. We demonstrated that none of the exposed Ae. aegypti herein became infected with BUNV via oral exposure, which in turn meant there was no opportunity for ToT. We did show some potential uptake of virus from contaminated larval habitat, but no subsequent transstadial transmission was observed. Thus, there is some potential for refraction to BUNV in Ae. aegypti populations as our population did not support infection. As seen with RVFV [34], other Ae. spp may be involved in this alternative BUNV transmission modality. Likely there are overlapping ecological factors that contribute to the similarities in the observed transmission of BUNV and RVFV, such as an overall suitability for multiple mosquito species in the region, including seasonal rainfall and other weather patterns that affect mosquito population dynamics in general. However, our data do not rule out the possibility of urbanization of BUNV in other Ae. aegypti populations, as another population of this species was found to be moderately competent [39].
These data could also be of use during surveillance efforts during a BUNV disease outbreak. The sensitivity assay demonstrates a high level of sensitivity for this primer set, as 9/10 samples with approximately ∼5 RNA copies per 10 mosquitoes in 1,000 uL and 100% of samples with ∼50 copies tested positive for BUNV via qRT-PCR, with a calculated LOD of 6.6 copies. Previous RT-PCR assays have low-end detections of BUNV RNA at relatively similar levels (700 copies and 2–20 copies per mL) [57, 58]. Further, it was previously found that in Ae. spp infected with Cache Valley Virus, an Orthobunyavirus in the Bunyamwera serogroup, the average titer of infectious virus ranged from approximately 3–5 logTCID50 [35]. This assay falls well within these limits and is competitive with other available assays, suggesting use as a surveillance tool is plausible.
In areas with RVF transmission, contact with blood and fluids from cattle is highly associated with seropositivity to RVFV [27, 59], indicating the importance of environmentally mediated transmission for the epidemiology of this virus. Our data does support the potential for environmentally mediated transmission of BUNV through blood and/or sera contamination, possibly from processing of meat or contaminated fomites/instruments. In higher volumes (up to 5 mL initial volume in tubes), infectiousness was retained for up to 28 days. At lower volumes over larger surface areas (plates), infectiousness was shorter owing to the shorter time to dried up substrate, but was still retained up to a week. The importance of identifying this potential novel transmission modality of BUNV lies in the potential for education and increased biosafety and infection control practices in at-risk communities, where PPE use is not high [60]. Recently, it was reported that fomite transmission might contribute to the movement of RVFV from rural areas to urban, as cattle may be moved to satiate the demand for meat in human dense areas [61]. This risk is shared by BUNV, as this data demonstrates this similarity in risk of environmentally mediated transmission between RVFV and BUNV. Further, the finding that dried samples were unlikely to harbor infectious virus speaks to the possibility for moisture control as a biosafety measure in areas where this risk exists.
Additionally, the stability of BUNV in whole blood at an environmentally relevant temperature may also inform surveillance efforts. Blood collected from suspected BUNV infected cattle that was unable to be immediately preserved via cooling may still be of use for qualitative surveillance purposes. This also suggests a potential use of blood that was collected indirectly or from meat processing facilities. Butchers and slaughterhouses as a point of surveillance may help inform on the epidemiology of an outbreak due to the concentration of animals to a central location [61]. Furthermore, environmental stability of BUNV in liquid may assist in less conventional surveillance methods for arboviruses, as the potential for wastewater surveillance of other arboviruses has been discussed [62].
While this experiment simulated field conditions in terms of temperature and humidity, additional factors may affect long-term BUNV stability and infectivity that we did not address herein. Both abiotic and biological contaminates found in the environment may alter the stability and infectivity by affecting the length of stability and/or probability of infectiousness over time. Furthermore, the physical environment may also influence BUNV stability, such as the moisture content of soil or rain influencing substrate moisture content which may extend the duration of infectiousness. While this is a limitation of our study, it only further emphasizes a need for a One Health and systems approach when characterizing understudied pathogens.
Given the role of animal and environmental factors and the potential to infect humans, viewing understudied Orthobunyaviruses such as BUNV through a One Health scope is immensely important, as changes animal, human, or the environmental conditions in the area may alter the epidemiology of BUNV. Thus, the data herein is an important addition to the knowledge of this neglected tropical disease and highlights the need for considering atypical transmission routes when cataloging the eco-epidemiology of viruses. It is important that we continue to investigate the interplay between these factors to be prepared to respond to the dynamic pressures of viral transmission and disease outbreaks.
Author contributions
Both authors were equally involved in hypothesis generation, experimental design, and writing and review of the manuscript. Experimentation was performed by ET. Data analysis performed by ET and overseen by RC. All authors contributed to the article and approved the submitted version.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Funding
This work was partially funded by the School of Veterinary Medicine and USDA 1433 Animal Health and Disease Research Program (FAIN/Award Number: NI21AHDRXXXXG064).
Acknowledgments
The authors would like to acknowledge Mr. Alex Stackhouse for his assistance in the course of the project and for Kameko the cat for her generous help as a paperweight.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
Please note that the review of this paper was conducted at the previous publisher, SAGE.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.ebm-journal.org/articles/10.3389/ebm.2024.10114/full#supplementary-material
References
1. Dutuze, MF, Ingabire, A, Gafarasi, I, Uwituze, S, Nzayirambaho, M, and Christofferson, RC. Identification of Bunyamwera and possible other orthobunyavirus infections and disease in cattle during a Rift Valley fever outbreak in Rwanda in 2018. Am J Trop Med Hyg (2020) 103:183–9. doi:10.4269/ajtmh.19-0596
2. Gerdes, GH. Rift valley fever. Vet Clin North America: Food Anim Pract (2002) 18:549–55. doi:10.1016/s0749-0720(02)00029-4
3. Peyre, M, Chevalier, V, Abdo-Salem, S, Velthuis, A, Antoine-Moussiaux, N, Thiry, E, et al. A systematic scoping study of the socio-economic impact of Rift Valley fever: research gaps and needs. Zoonoses and Public Health (2015) 62:309–25. doi:10.1111/zph.12153
4. Siam, AL, Meegan, JM, and Gharbawi, KF. Rift Valley fever ocular manifestations: observations during the 1977 epidemic in Egypt. Br J Ophthalmol (1980) 64:366–74. doi:10.1136/bjo.64.5.366
5. Daubney, R, Hudson, JR, and Garnham, PC. Enzootic hepatitis or rift valley fever. An undescribed virus disease of sheep cattle and man from east africa. J Pathol Bacteriol (1931) 34:545–79. doi:10.1002/path.1700340418
6. Samy, AM, Peterson, AT, and Hall, M. Phylogeography of Rift Valley fever virus in africa and the arabian peninsula. PLOS Negl Trop Dis (2017) 11:e0005226. doi:10.1371/journal.pntd.0005226
7. Davies, FG, Linthicum, KJ, and James, AD. Rainfall and epizootic Rift Valley fever. Bull World Health Organ (1985) 63:941–3.
8. Dutuze, MF, Nzayirambaho, M, Mores, CN, and Christofferson, RC. A review of Bunyamwera, batai, and ngari viruses: understudied orthobunyaviruses with potential one health implications. Front Vet Sci (2018) 5:69. doi:10.3389/fvets.2018.00069
10. Elliott, RM. Orthobunyaviruses: recent genetic and structural insights. Nat Rev Microbiol (2014) 12:673–85. doi:10.1038/nrmicro3332
11. Kuhn, JH, Abe, J, Adkins, S, Alkhovsky, SV, Avšič-Županc, T, Ayllón, MA, et al. Annual (2023) taxonomic update of RNA-directed RNA polymerase-encoding negative-sense RNA viruses (realm Riboviria: kingdom Orthornavirae: phylum Negarnaviricota). J Gen Virol (2023) 104:001864. doi:10.1099/jgv.0.001864
12. Sasaya, T, Palacios, G, Briese, T, Di Serio, F, Groschup, MH, Neriya, Y, et al. ICTV virus taxonomy profile: phenuiviridae 2023. J Gen Virol (2023) 104. doi:10.1099/jgv.0.001893
13. Hughes, HR, Adkins, S, Alkhovskiy, S, Beer, M, Blair, C, Calisher, CH, et al. ICTV virus taxonomy profile: peribunyaviridae. J Gen Virol (2020) 101:1–2. doi:10.1099/jgv.0.001365
14. Nashed, NW, Olson, JG, and El-Tigani, A. Isolation of batai virus (Bunyaviridae:Bunyavirus) from the blood of suspected malaria patients in Sudan. Am J Trop Med Hyg (1993) 48:676–81. doi:10.4269/ajtmh.1993.48.676
15. Waddell, L, Pachal, N, Mascarenhas, M, Greig, J, Harding, S, Young, I, et al. Cache Valley virus: a scoping review of the global evidence. Zoonoses and Public Health (2019) 66:739–58. doi:10.1111/zph.12621
16. Hofmann, M, Wiethölter, A, Blaha, I, Jöst, H, Heinemann, P, Lehmann, M, et al. Surveillance of Batai virus in bovines from Germany. Clin Vaccin Immunol (2015) 22:672–3. doi:10.1128/cvi.00082-15
17. Tauro, LB, Rivarola, ME, Lucca, E, Mariño, B, Mazzini, R, Cardoso, JF, et al. First isolation of Bunyamwera virus (Bunyaviridae family) from horses with neurological disease and an abortion in Argentina. Vet J (2015) 206:111–4. doi:10.1016/j.tvjl.2015.06.013
18. Yanase, T, Kato, T, Yamakawa, M, Takayoshi, K, Nakamura, K, Kokuba, T, et al. Genetic characterization of Batai virus indicates a genomic reassortment between orthobunyaviruses in nature. Arch Virol (2006) 151:2253–60. doi:10.1007/s00705-006-0808-x
19. Linthicum, KJ, Davies, FG, Kairo, A, and Bailey, CL. Rift Valley fever virus (family Bunyaviridae, genus Phlebovirus). Isolations from Diptera collected during an inter-epizootic period in Kenya. J Hyg (1985) 95:197–209. doi:10.1017/s0022172400062434
20. Adeniji, JA. Vector competence of NIGERIAN strain of Aedes aegypti linn. (DIPTERA: CULICIDAE) for Rift Valley fever virus. Department of Virology, Faculty of Basic Medical Sciences. University of Ibadan (2002). Available from: https://library.adhl.africa/handle/123456789/12263 (Accessed November 20, 2023).
21. Mohamed, R, Abdelgadir, DM, and Bashab, H. Transovarian transmission of Rift Valley fever virus by two species of mosquitoes in Khartoum state (Sudan): aedes vexans (Meigen) and culex quinquefasciatus (Say). Sudan J Public Health (2013) 8:164–70.
22. Turell, MJ, Reeves, WC, and Hardy, JL. Transovarial and trans-stadial transmission of California encephalitis virus in Aedes dorsalis and Aedes melanimon. Am J Trop Med Hyg (1982) 31:1021–9. doi:10.4269/ajtmh.1982.31.1021
23. Vargas, HCM, Farnesi, LC, Martins, AJ, Valle, D, and Rezende, GL. Serosal cuticle formation and distinct degrees of desiccation resistance in embryos of the mosquito vectors Aedes aegypti, Anopheles aquasalis and Culex quinquefasciatus. J Insect Physiol (2014) 62:54–60. doi:10.1016/j.jinsphys.2014.02.001
24. Wright, D, Kortekaas, J, Bowden, TA, and Warimwe, GM. Rift Valley fever: biology and epidemiology. J Gen Virol (2019) 100:1187–99. doi:10.1099/jgv.0.001296
25. Valdez, LD, Sibona, GJ, Diaz, LA, Contigiani, MS, and Condat, CA. Effects of rainfall on Culex mosquito population dynamics. J Theor Biol (2017) 421:28–38. doi:10.1016/j.jtbi.2017.03.024
26. Ha, TV, Kim, W, Nguyen-Tien, T, Lindahl, J, Nguyen-Viet, H, Thi, NQ, et al. Spatial distribution of Culex mosquito abundance and associated risk factors in Hanoi, Vietnam. PLOS Negl Trop Dis (2021) 15:e0009497. doi:10.1371/journal.pntd.0009497
27. Nyakarahuka, L, de St Maurice, A, Purpura, L, Ervin, E, Balinandi, S, Tumusiime, A, et al. Prevalence and risk factors of Rift Valley fever in humans and animals from Kabale district in Southwestern Uganda, 2016. PLOS Negl Trop Dis (2018) 12:e0006412. doi:10.1371/journal.pntd.0006412
28. Gerken, KN, Labeaud, AD, Mandi, H, L’Azou Jackson, M, Breugelmans, JG, and King, CH. Paving the way for human vaccination against Rift Valley fever virus: a systematic literature review of RVFV epidemiology from 1999 to 2021. PLOS Negl Trop Dis (2022) 16:e0009852. doi:10.1371/journal.pntd.0009852
29. Nicholas, DE, Jacobsen, KH, and Waters, NM. Risk factors associated with human Rift Valley fever infection: systematic review and meta-analysis. Trop Med Int Health (2014) 19:1420–9. doi:10.1111/tmi.12385
30. Craig, DE, Thomas, WJ, and Desanctis, AN. Stability of Rift Valley fever virus at 4 C. Appl Microbiol (1967) 15:446–7. doi:10.1128/am.15.2.446-447.1967
32. European Food Safety Authority EFSA. Opinion of the scientific panel on animal health and welfare (AHAW) on a request from the commission related to “the risk of a Rift Valley fever incursion and its persistence within the community”. EFSA J (2005) 3:238. doi:10.2903/j.efsa.2005.238
33. Francis, T, and Magill, TP. Rift Valley Fever. J Exp Med (1935) 62:433–48. doi:10.1084/jem.62.3.433
34. Turell, MJ, Beaman, JR, and Linthicum, KJ. Transmission of Rift Valley fever virus by adult mosquitoes after ingestion of virus as larvae. Am J Trop Med Hyg (1990) 43:677–80. doi:10.4269/ajtmh.1990.43.677
35. Ayers, VB, Huang, Y-JS, Lyons, AC, Park, SL, Dunlop, JI, Unlu, I, et al. Infection and transmission of Cache Valley virus by Aedes albopictus and Aedes aegypti mosquitoes. Parasites and Vectors (2019) 12:384. doi:10.1186/s13071-019-3643-0
36. Chan, KK, Auguste, AJ, Brewster, CC, and Paulson, SL. Vector competence of Virginia mosquitoes for zika and Cache Valley viruses. Parasites and Vectors (2020) 13:188. doi:10.1186/s13071-020-04042-0
37. EFSA Panel on Animal Health and Welfare AHAW. Scientific opinion on Rift Valley fever. EFSA J (2013) 11:3180. doi:10.2903/j.efsa.2013.3180
38. Mweya, CN, Kimera, SI, Kija, JB, and Mboera, LEG. Predicting distribution of Aedes aegypti and Culex pipiens complex, potential vectors of rift valley fever virus in relation to disease epidemics in East Africa. Infect Ecol Epidemiol (2013) 3:21748. doi:10.3402/iee.v3i0.21748
39. Odhiambo, C, Venter, M, Chepkorir, E, Mbaika, S, Lutomiah, J, Swanepoel, R, et al. Vector competence of selected mosquito species in kenya for ngari and bunyamwera viruses. J Med Entomol (2014) 51:1248–53. doi:10.1603/me14063
40. Kading, RC, Crabtree, MB, Bird, BH, Nichol, ST, Erickson, BR, Horiuchi, K, et al. Deletion of the NSm virulence gene of rift valley fever virus inhibits virus replication in and dissemination from the midgut of Aedes aegypti mosquitoes. Plos Negl Trop Dis (2014) 8:e2670. doi:10.1371/journal.pntd.0002670
41. Seufi, AM, and Galal, FH. Role of culex and anopheles mosquito species as potential vectors of rift valley fever virus in Sudan outbreak, 2007. BMC Infect Dis (2010) 10:65. doi:10.1186/1471-2334-10-65
42. Hughes, MT, Gonzalez, JA, Reagan, KL, Blair, CD, and Beaty, BJ. Comparative potential of Aedes triseriatus, Aedes albopictus, and Aedes aegypti (Diptera: Culicidae) to transovarially transmit La crosse virus. J Med Entomol (2006) 43:757–61. doi:10.1093/jmedent/43.4.757
43. Dutuze, MF, Mayton, EH, Macaluso, JD, and Christofferson, RC. Comparative characterization of the reassortant Orthobunyavirus Ngari with putative parental viruses, Bunyamwera and Batai: in vitro characterization and ex vivo stability. J Gen Virol (2021) 102:001523. doi:10.1099/jgv.0.001523
44. Habiyaremye, G, Jairu, N, De la Paix Mupenzi, J, Ngamije, J, Baragahoranye, I, and Karangwa, A. Statistical analysis of climatic variables and prediction outlook in Rwanda. East Afr J Sci Technol (2012) 1:27–34.
45. Mayton, EH, Tramonte, AR, Wearing, HJ, and Christofferson, RC. Age-structured vectorial capacity reveals timing, not magnitude of within-mosquito dynamics is critical for arbovirus fitness assessment. Parasites and Vectors (2020) 13:310. doi:10.1186/s13071-020-04181-4
46. Schmidt, OW, Cooney, MK, and Kenny, GE. Plaque assay and improved yield of human coronaviruses in a human rhabdomyosarcoma cell line. J Clin Microbiol (1979) 9:722–8. doi:10.1128/jcm.9.6.722-728.1979
47. Ragan, I, Hartson, L, Pidcoke, H, Bowen, R, and Goodrich, R. Pathogen reduction of SARS-CoV-2 virus in plasma and whole blood using riboflavin and UV light. PLOS ONE (2020) 15:e0233947. doi:10.1371/journal.pone.0233947
48. Mayton, EH, Hernandez, HM, Vitek, CJ, and Christofferson, RC. A method for repeated, longitudinal sampling of individual Aedes aegypti for transmission potential of arboviruses. Insects (2021) 12:292. doi:10.3390/insects12040292
49. Molina-Cruz, A, Bennett, K, Barillas-Mury, C, Richardson, J, Black, W, and Gupta, L. Effect of mosquito midgut trypsin activity on dengue-2 virus infection and dissemination in Aedes aegypti. Am J Trop Med Hyg (2005) 72:631–7. doi:10.4269/ajtmh.2005.72.631
50. Veronesi, E, Paslaru, A, Ettlin, J, Ravasi, D, Flacio, E, Tanadini, M, et al. Estimating the impact of consecutive blood meals on vector competence of Aedes albopictus for chikungunya virus. Pathogens (2023) 12:849. doi:10.3390/pathogens12060849
51. Armstrong, PM, Ehrlich, HY, Magalhaes, T, Miller, MR, Conway, PJ, Bransfield, A, et al. Successive blood meals enhance virus dissemination within mosquitoes and increase transmission potential. Nat Microbiol (2019) 5:239–47. doi:10.1038/s41564-019-0619-y
52. Merkes, CMKK, Allison, MJ, Goldberg, C, Helbing, CC, Hunter, ME, Jackson, CA, et al. Generic qPCR limit of detection (LOD)/limit of quantification (LOQ) calculator (2019). Available from: https://github.com/cmerkes/qPCR_LOD_Calc.2019 (Accessed November 20, 2023).
53. Ritz, C, Baty, F, Streibig, JC, and Gerhard, D. Dose-response analysis using R. PLOS ONE (2015) 10:e0146021. doi:10.1371/journal.pone.0146021
54. Xu, S, Chen, M, Feng, T, Zhan, L, Zhou, L, and Yu, G. Use ggbreak to effectively utilize plotting space to deal with large datasets and outliers. Front Genet (2021) 12:774846. doi:10.3389/fgene.2021.774846
55. Gu, W, Lampman, R, and Novak, RJ. Assessment of arbovirus vector infection rates using variable size pooling. Med Vet Entomol (2004) 18:200–4. doi:10.1111/j.0269-283x.2004.00482.x
56. Bergren, N, and Kading, R. The ecological significance and implications of transovarial transmission among the vector-borne bunyaviruses: a review. Insects (2018) 9:173. doi:10.3390/insects9040173
57. Villinger, J, Mbaya, MK, Ouso, D, Kipanga, PN, Lutomiah, J, and Masiga, DK. Arbovirus and insect-specific virus discovery in Kenya by novel six genera multiplex high-resolution melting analysis. Mol Ecol Resour (2017) 17:466–80. doi:10.1111/1755-0998.12584
58. Lambert, AJ, and Lanciotti, RS. Consensus amplification and novel multiplex sequencing method for S segment species identification of 47 viruses of the orthobunyavirus, phlebovirus, and nairovirus genera of the family bunyaviridae. J Clin Microbiol (2009) 47:2398–404. doi:10.1128/jcm.00182-09
59. Nicholas, DE, Jacobsen, KH, and Waters, NM. Risk factors associated with human Rift Valley fever infection: systematic review and meta-analysis. Trop Med Int Health (2014) 19:1420–9. doi:10.1111/tmi.12385
60. Smith, LJ, Schurer, JM, Ntakiyisumba, E, Shyaka, A, and Amuguni, JH. Rift Valley fever knowledge, mitigation strategies and communication preferences among male and female livestock farmers in Eastern Province, Rwanda. PLOS Negl Trop Dis (2021) 15:e0009705. doi:10.1371/journal.pntd.0009705
61. Gerken, KN, Ndenga, BA, Owuor, KO, Winter, CA, Seetah, K, and LaBeaud, AD. Leveraging livestock movements to urban slaughterhouses for wide-spread Rift Valley fever virus surveillance in Western Kenya. One Health (2022) 15:100457. doi:10.1016/j.onehlt.2022.100457
Keywords: Orthobunyavirus, Bunyamwera, Aedes aegypti, environmental transmission, transovarial transmission
Citation: Turner EA and Christofferson RC (2024) Exploring the transmission modalities of Bunyamwera virus. Exp. Biol. Med. 249:10114. doi: 10.3389/ebm.2024.10114
Received: 07 August 2023; Accepted: 11 December 2023;
Published: 15 February 2024.
Copyright © 2024 Turner and Christofferson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebecca C. Christofferson, rcarri1@lsu.edu
 Erik A. Turner
Erik A. Turner Rebecca C. Christofferson
Rebecca C. Christofferson