Abstract
Investigations on some innate immunity proteins can yield misleading information, as investigators often rely on static measurements and assume a direct correlation to function. As protein function is often not directly proportional to protein abundance, and mechanistic pathways are interconnected and under constant feedback regulatory control, functional analysis is required. In this study, we used functional mass spectrometry to measure anti-protease and complement activity in plasma obtained from coronavirus disease 2019 (COVID-19) patients. Our data suggests that within 48 h of hospital admission, COVID-19 patients undergo a protease storm with significantly elevated neutrophil elastase (p < 0.001) and lymphocyte granzyme B (p < 0.01), while, anti-protease activity is significantly increased, including alpha-1 antitrypsin (AAT; p < 0.001) and alpha-1-antichymotrypsin (ACT; p < 0.001). Concurrently, the ratio of C3a to C3beta activity significantly decreased with increasing COVID-19 severity, suggesting more complement activation (Mild COVID-19 p < 0.05; Severe COVID-19 p < 0.001). Activity levels of AAT, ACT and C3a/C3beta remained unchanged over 10 hospital days. Our data suggests that COVID-19 is associated with both a protease storm and complement activation, with the former somewhat balanced with increased anti-protease activity. Evaluation of the AAT/ACT ratio and C3a/C3beta ratio indicated that COVID-19 severity is associated with both neutrophil elastase neutralization and complement activation.
Impact statement
Our work is important as many investigations on innate immunity proteins can yield misleading information, as investigators often rely on static measurements. We used a novel approach which relies on functional mass spectrometry to measure the active site, thereby inferring function. We show that with increasing COVID-19 severity the anti-protease activity is significantly increased and that the ratio of C3a to C3beta activity significantly decreased. Our data is novel in that it shows COVID-19 severity is associated with both neutrophil elastase neutralization, together with complement activation.
Introduction
Coronavirus disease 2019 (COVID-19) is associated with a strong innate immune response [1], mediated by neutrophils, lymphocytes, and the complement system. Severe COVID-19 involves the acute replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that results in a strong innate inflammatory response associated with damage to the airways, hyperactivation of the complement system, and multiorgan failure [2–4]. This innate immune response is mediated by monocytes, macrophages and neutrophils and an elevation in serum chemokines.
Neutrophil elastase and granzyme B were previously identified as significantly elevated in severe COVID-19 [5]. Similarly, other studies have shown an upregulation of neutrophil degranulation proteins in SARS-CoV-2 positive nasal swabs [6] and that neutrophil serine proteases are key mediators in the innate and adaptive immune functions of immune cells [7] and inflammatory disease [8]. Dysregulated proteolysis is central to inflammatory disease and examining the anti-protease and complement responses could provide a better understanding of the disease process and aid the development of targeted therapies.
The anti-protease activities of serine protease inhibitors (serpins) have been identified as potential clinical biomarkers of COVID-19 severity [9]. Proteomic methods routinely use immunoassays and ligand binding assays to measure protein abundance, but these methods are designed to quantify total protein levels and not protein activity. Studying serpins poses a proteomic challenge as structural information is required to distinguish between active and inactive forms.
Quantifying protein activity would provide a more accurate measure to investigate the function of these factors in the innate immune response in COVID-19 [10, 11]. Mass spectrometry enables functional proteomics to inform on protein structure and functional status for select proteins. Additionally, mass spectrometry is a powerful proteomics tool because it requires small sample sizes, is quantitative, and is amenable to customizable sample preparation and enrichment techniques.
In this study, we aimed to examine differences in serum levels of neutrophil elastase and granzyme B proteases, and to examine the anti-protease activity of their associated serpins, alpha-1-antitrypsin (ATT) and alpha-1-antichymotrypsin (ACT) as well as complement activation activity of factor 3 (C3) and complement factor 3 a (C3a) in context of COVID-19 severity. We present a functional mass spectrometry approach that allows active protein to be distinguished from total protein for further investigation of these potential COVID-19 biomarkers.
Materials and methods
Study participants and clinical data
Patients were enrolled March-June 2000 after admission to our participating academic hospital, London Health Sciences Centre (London, Ontario). COVID-19 was first suspected based on standard hospital screening procedures, and then confirmed as part of standard hospital testing by detection of two SARS-CoV-2 viral genes using polymerase chain reaction (Dual target assay, specific target unique to SARS-Cov-2 along with a conserved region of the E-gene; Roche cobas® SARS-COV-2 Test). Patient baseline characteristics were recorded on admission and included age, sex, comorbidities, medications, hematologic labs, creatinine, arterial partial pressure to inspired oxygen (P/F) ratio, and chest x-ray findings. We calculated Multiple Organ Dysfunction Score (MODS) [12] and Sequential Organ Failure Assessment (SOFA) [13] score to illustrate their illness severity. We also recorded clinical interventions received during the observation period including use of antibiotics, anti-viral agents, systemic corticosteroids, vasoactive medications, venous thromboembolism (VTE) prophylaxis, anti-platelet or anti-coagulation treatment, renal replacement therapy, high flow oxygen therapy, and mechanical ventilation (invasive and non-invasive). For comparison to critically ill patients, we also included an age- and sex-matched non-critically ill COVID-19 patient cohort who were admitted to the hospital respiratory ward with moderate disease. All COVID-19 patients were non-vaccinated for SARS-CoV-2. Final participant groups contained were constructed by age- and sex-matching patient cohorts with healthy controls without disease, acute illness, or prescription medications (collected prior to the SARS-CoV-2 pandemic). Each of the three groups consisted of 15 participants.
Blood draws
Standard operating procedures were used to ensure all samples were treated rapidly and equally (available at1). The first blood sample was obtained within 48 h of hospital admission via indwelling catheters using vacuum serum separator tubes and placed immediately on ice. For ICU patients, additional blood samples were drawn at 4, 7, and 10 days. If venipuncture was required, research blood draws were coordinated with a clinically indicated blood draw. In keeping with accepted research phlebotomy protocols for adult patients, blood draws did not exceed maximal volumes. Once transferred to a negative pressure hood, blood was centrifuged and sera isolated, aliquoted at 250 µL and frozen at −80°C. All samples remained frozen until use and freeze/thaw cycles were avoided.
Protease measurements
The levels of serum neutrophil elastase and granzyme B, were determined using multiplexed biomarker immunoassay kits according to manufacturers’ instructions (MilliporeSigma, 400 Summit Drive, Burlington, MA), as we have previously described [5]. Analytes were measured using a Bio-PlexTM 200 Suspension Array system (Bio-Rad Laboratories, Hercules, CA), which used Luminex xMAPTM fluorescent bead-based technology (Luminex Corp, Austin, TX). Bioanalyte concentrations were calculated from standard curves using five-parameter logistic regression in Bio-Plex Manager 6.1 software. Serum concentrations are reported as pg/mL.
Albumin and IgG depletion
A 60 mg aliquot of AlbuSorb™ PLUS beads (Biotech Support Group, Monmouth Junction NJ) was conditioned with 400 µL of Binding buffer (BB1), then the liquid was removed through a spin-filter by centrifugation for 2 min at 1000 × g, and repeated. A 25 µL aliquot of serum was diluted with 250 µL BB1 buffer, and clarified by passing through another spin-filter at 9,000 × g for 1 min. The clarified serum was loaded onto the conditioned AlbuSorb™ PLUS beads and incubated on a rotator at room temperature for 15 min then centrifuged for 4 min at 9,000 × g to collect the filtrate for further processing.
Reduction and digestion
After depletion, an equal volume of serum filtrate ( ̴20 µg) was loaded onto SDS-PAGE as gel plug, and in-gel digested with a standard protocol; proteins in the gel bands were reduced with 10 mM dithiothreitol (DTT) for 30 min at 60°C, alkylated with 20 mM iodoacetamide for 45 min at room temperature in the dark, and digested overnight with 0.4 µg of trypsin (Pierce MS Grade (ThermoFisher) at 37°C. Peptides were extracted twice with 5% formic acid, 60% acetonitrile and dried under vacuum.
Target peptide selection
The target peptides were selected to report specific regions of the protein(s) that infer functionality (Table 1). For Complement C3, activation requires the proteolytic cleavage of the region C3a from the rest of the protein, generating Activated C3b. Two peptides were chosen from sequences of the C3 beta chain region – part of both the intact C3 and the Activated C3b subform (the C3 beta chain signal value was the average of the two peptides). One peptide was chosen from C3a sequence: the region proteolytically cleaved upon C3 Convertase activation [14]. As this method only considers observations at the protein level, less C3a relative to C3 beta would be indicative of proportionately more activated C3b subforms. For both Serpins (AAT and ACT), tryptic peptides which span the Reactive Center Loop (RCL) region were chosen, as these regions represent the proteins’ potential function as an inhibitor, any loss of which would negate inhibitory potential [14]. Taken together, the peptides selected from these regions thus can serve as surrogates for reporting functional activity.
TABLE 1
| Protein | Target peptide sequence |
|---|---|
| Complement C3a region | FISLGEAC [+57]K |
| Complement C3 beta chain region | FVTVQATFGTQVVEK |
| Complement C3 beta chain region | IPIEDGSGEVVLSR |
| Alpha-1-Antitrypsin RCL Spanning Intact Region | GTEAAGAMFLEAIPMSIPPEVK |
| Alpha-1-Antichymotrypsin RCL Spanning Intact Region | ITLLSALVETR |
PRM target peptides for LC-MS/MS.
LC-MS/MS
0.2 µg of system-independent retention time (iRT) peptides (Biognosys) were added to each digested sample and 1% of the sample was analyzed by LC-MS/MS using a Dionex RSLC nano system coupled to an Orbitrap Eclipse Tribrid mass spectrometer (ThermoFisher). The peptides were loaded onto a fused silica trap column (Acclaim PepMap 100, 75 µm × 2 cm, ThermoFisher). After washing for 5 min at 5 μL/min with 0.1% trifluoroacetic acid (TFA), the trap column was brought in-line with an analytical column (Nanoease MZ peptide BEH C18, 130A, 1.7 µm, 75 µm × 250 mm, Waters) for LC-MS/MS. Peptides were fractionated at 300 nL/min using a segmented linear gradient 4–15% B in 5 min (where A: 0.2% formic acid, and B: 0.16% formic acid, 80% acetonitrile), 15–50% B in 50 min, and 50–90% B in 15 min. Parallel Reaction Monitoring (PRM) method template was used to target selected ions on the target list throughout the run (10–60 min). The target list is shown in Table 1. The isolation width was set at 1.6 Da. Automatic Gain Control (AGC) was set at 2E5 and max ion time set at 150 ms. Ions were fragmented by Higher Energy Collision Dissociation (HCD) using 30% relative collision energy and scanned in the Orbitrap with a resolution of 30,000 m/z. The data was analyzed using Skyline-daily (beta) [15] with a spectral library generated previously. Only peptides with dotp > 0.8 were accepted. All fragmented ions from the mass spectrum that corresponded to the target peptides, generate an accumulated spectral intensity. Relative spectral intensity data is normalized to the average value of the iRT peptide ions for each individual run. Complement C3 beta was reported as an average value of the two C3 beta peptides.
Population statistics
Medians (IQRs) and frequencies (%) were used to report patient baseline characteristics for continuous and categorical variables, respectively; continuous variables were compared using Mann-Whitney U tests (and Kruskal-Wallis tests, as appropriate), and categorical variables were compared using Fisher’s exact chi-square tests. p-values <0.05 (*) were considered statistically significant. All analyses were conducted using GraphPad Prism version 9.2.0 for Windows (GraphPad Software, California, United States) and SPSS version 27 (IBM Corp., Armonk, NY, United States).
Results
Participant demographics and clinical variables
We investigated a total of 45 age- and sex-matched subjects, including 15 severe COVID-19 ICU patients, 15 mild COVID-19 ward patients and 15 healthy subjects. The mean age and sex distribution were similar between groups. Subject demographics and clinical variables are presented in Table 2. COVID-19 patients were likely infected with wild-type SARS-CoV-2 (B.1 strain) given the dates of recruitment and sample collection.
TABLE 2
| Clinical variable | Healthy subjects | Mild COVID-19 | Severe COVID-19 |
|---|---|---|---|
| Age in years, median (IQR) | 61 (46–66) | 59 (46–68) | 60 (47–67) |
| Sex, female: male | 7:8 | 7:8 | 7:8 |
| Comorbidities, n (%) | |||
| Diabetes Hypertension Coronary Artery Disease Heart Failure Chronic Kidney Disease Cancer COPD |
3 (20.0) 6 (40.0) 0 (0) 0 (0) 1 (6.6) 1 (6.6) 0 (0) |
3 (20.0) 5 (33.3) 0 (0) 0 (0) 0 (0) 1 (6.6) 0 (0) |
|
| MODS, median (IQR) | - | 4.5 (4.0–5.0) | |
| SOFA, median (IQR) | - | 6.0 (2.0–7.0) | |
| Presenting Symptoms, n (%) | |||
| Fever Cough Anosmia/ageusia Pharyngitis Headache Myalgias Dyspnea Chest Pain Nausea/vomiting/diarrhea |
11 (73.3) 12 (80.0) 3 (20.0) 3 (20.0) 2 (13.3) 9 (60.0) 13 (86.6) 0 (0) 6 (40.0) |
- - - - - - - - - |
|
| Laboratory, median (IQR) Hemoglobin White Blood Cell Count Neutrophils Lymphocytes Platelets Creatinine INR Lactate CRP D-Dimer Albumin |
137.0 (117.0–142.0) 7.5 (4.7–10.4) 6.6 (3.5–8.4) 1.0 (0.5–1.2) 236.0 (187.0–277.0) 70.0 (54.0–92.0) 1.0 (1.0–1.1) 1.5 (1.3–2.2) 90.6 (68.1–112.1) 616.0 (441.0–3,367.0) 34.0 (31.5–36.5) |
121.0 (104.0–136.0) 8.2 (6.9–11.5) 7.3 (5.5–9.7) 0.7 (0.4–1.0) 210.0 (186.0–210.0) 81.0 (56.0–184.0) 1.2 (1.1–1.3) 1.2 (1.0–1.9) 133.9 (71.1–240.5) 3,964.0 (1224.0–14725.0) 30.0 (26.0–32.0) |
|
| Pulmonary Abnormalities, n (%) | |||
| Bilateral Infiltrates Unilateral Infiltrates Normal |
14 (93.3) 0 (0) 1 (6.6) |
15 (100) 0 (0) 0 (0) |
|
| P/F Ratio, median (IQR) | - | 93.0 (66.0–131.0) | |
| Interventions, n (%) | |||
| Antibiotics Steroids Remdesivir Tocilizumab Vasoactive medications High Flow Nasal Cannula Non-invasive MV Invasive MV Renal Replacement Therapy |
14 (93.3) 15 (100) 4 (26.6) 1 (6.6) 2 (13.3) 7 (46.6) 1 (6.6) 2 (13.3) 0 (0) |
15 (100) 10 (66.6) 0 (0) 0 (0) 14 (93.3) 10 (66.6) 4 (26.6) 14 (93.3) 3 (20.0) |
|
| ICU Days, median (IQR) | - | 17.0 (10.0–26.0) | |
| Hospital Days, median (IQR) | 8.0 (6.0–16.0) | - | |
| Outcome, n (%) | |||
| Alive Dead |
15 (100) 0 (0) |
8 (53.3) 7 (46.6) |
|
Subject demographics and clinical variables.
Serine protease measurements
We observed increased levels of neutrophil elastase and granzyme B in patients hospitalized for severe COVID-19. Immunoassay quantification of these proteases in serum indicate that within 48 h of admission, severe COVID-19 patients had significantly higher levels of neutrophil elastase (median 71.8 pg/mL versus 2.8 pg/mL; p > 0.001) (Figure 1A) and granzyme B (median 8.0 pg/mL versus 2.3 pg/mL; p = 0.006) (Figure 1B).
FIGURE 1
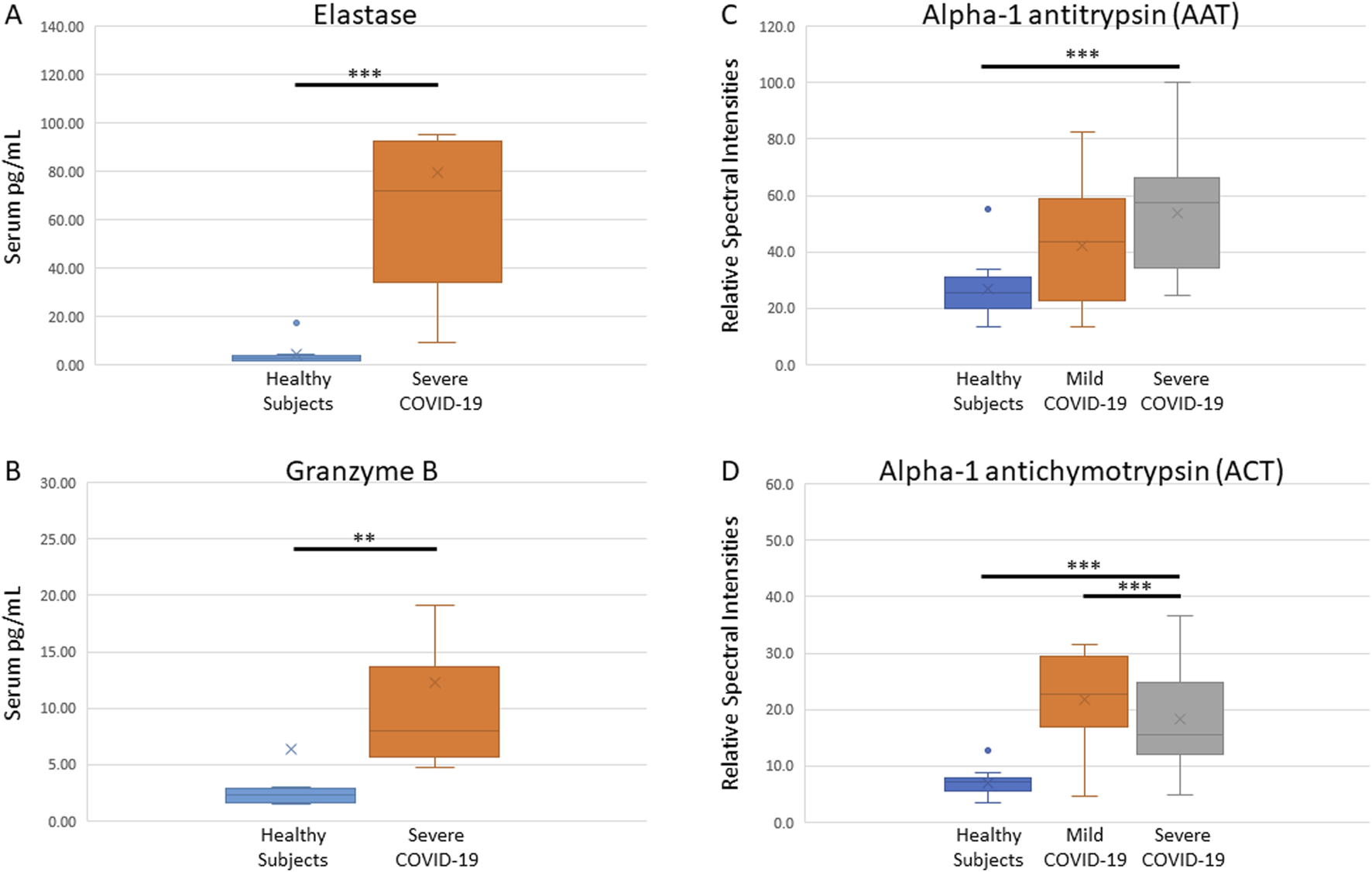
Measurements of serine proteases and functional analyses of their anti-proteases. Immunoassays show elevated levels of neutrophil elastase (A) and granzyme B (B), and functional proteomic analysis indicate high activity of AAT (C) and ACT (D) anti-proteases, in serum of COVID-19 infected patients within 48 h of hospital admission.
Anti-protease activity
Functional mass spectrometry was used to quantify the level of active serpins AAT and ACT in serum. We observed that within 48 h of hospital admission, anti-protease activity in COVID-19 patient serum is significantly increased. AAT activity was highest in severe COVID-19 patients (median 57.3, p < 0.001; Figure 1C) and ACT activity (median 22.6, p < 0.001; Figure 1D) was significantly higher in mild compared to severe cases. These findings indicate that compared to healthy subjects, AAT may be correlated with COVID-19 disease severity. Interestingly, ACT activity was elevated in mild COVID-19 but decreased in severe COVID-19 serum (median 15.6, p < 0.001). Although ACT is significantly elevated in serum of both the mild and severe COVID-19 population, ACT may be relatively depleted in serum with a greater protease load. Neither AAT nor ACT changed significantly over 10 days of ICU admission (AAT, p = 0.082; ACT p = 0.059).
Complement activation
We observed that serum Complement activation was associated with COVID-19 severity within 48 h of hospital admission, as inversely measured by the C3a/C3beta ratio (median 0.12, p < 0.001) (Figure 2A). The C3a/C3beta ratio did not change significantly over 10 days of ICU admission (p = 0.447; data not shown).
FIGURE 2
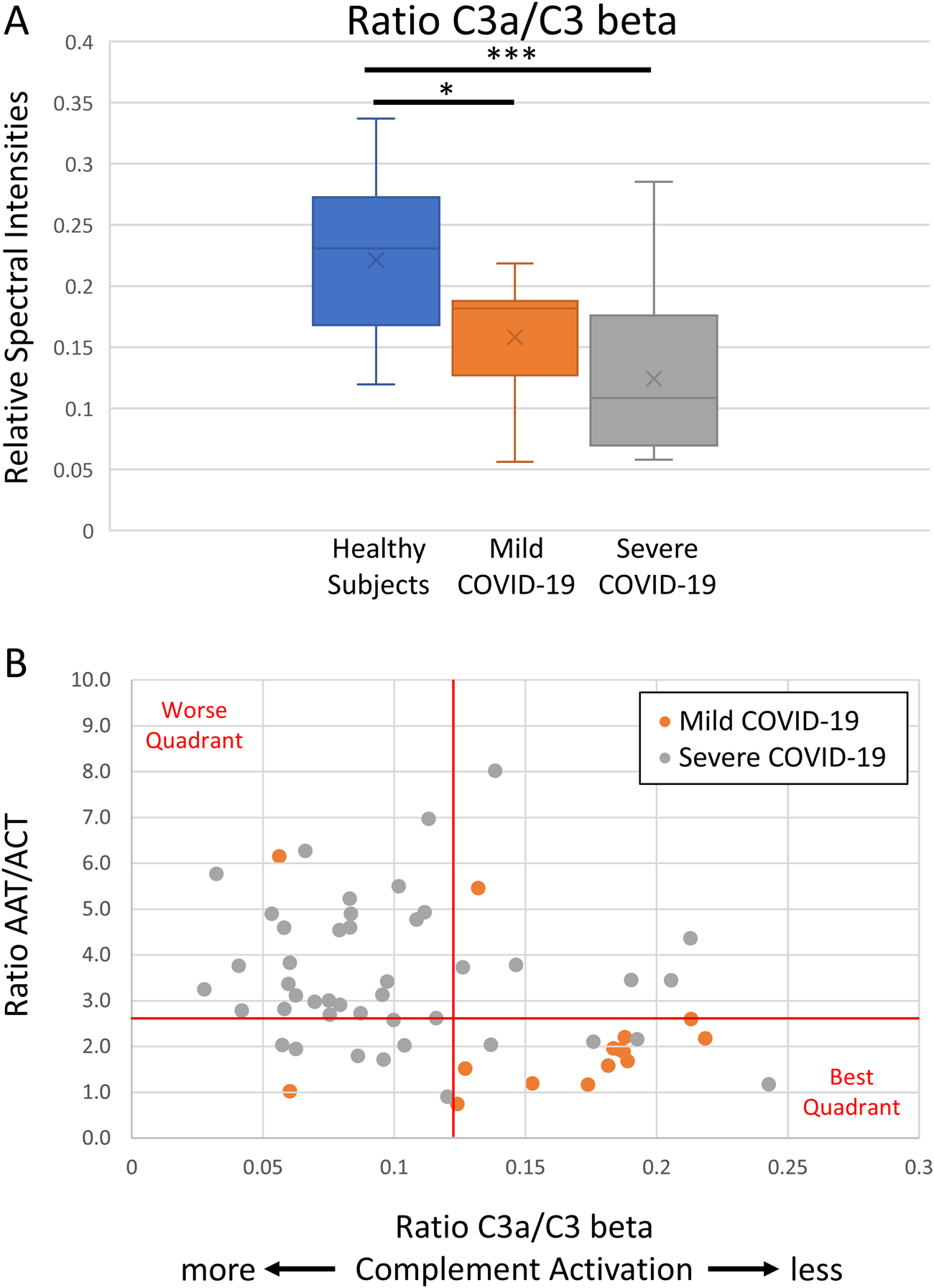
Functional analyses of complement activity and its role in COVID-19 severity. Functional proteomic analysis of serum complement activity indicates a decrease in C3a/C3 beta ratio (A), which is a metric of relative C3 activation, and a comparison of anti-protease activity to complement activation indicates milder COVID-19 cases cluster toward low anti-protease activity ratio (AAT/ACT; the ratio of the RCL tryptic peptides) and less complement activation (C3a/C3 beta) (B). The graphical line thresholds were chosen arbitrarily as an indicator for quadrant stratification. Severe COVID-19 data points were calculated from all ICU days.
Consequences of anti-protease activity versus complement activation
We compared anti-protease activity ratio (AAT/ACT) to complement activation (C3a/C3 beta) in both mild and severe COVID-19 cases. Our results indicate that milder COVID-19 cases cluster towards the lower right quadrant and associated with low anti-protease activity ratio and low complement activation (Figure 2B).
Discussion
In this study, we investigated two serine proteases, previously identified as elevated in the serum of COVID-19 ICU patients [5], to assess their functional activity using mass spectrometry. We compared healthy controls to COVID-19 patients with mild or severe disease. Our results showed increased anti-protease activity with disease severity and decreased complement activity in COVID-19 patients.
Participants were classified as mild or severe based on PCR results and symptom severity, all infected with wild-type SARS-CoV-2. Blood samples were collected within 48 h of hospital admission. While all mild patients survived, 47% of severe patients died. Healthy controls were obtained prior to the pandemic.
Immunoassays showed significantly higher levels of neutrophil elastase and granzyme B in severe COVID-19 patients within 48 h of admission, compared to healthy controls. These proteases are crucial for immune regulation, but excessive activity can cause tissue damage, cytotoxicity, and inflammation [7, 16]. Protease-induced degradation of the extracellular matrix also damages blood vessels, increasing vascular permeability and causing hypotension, often necessitating vasoactive support. The elevated neutrophil elastase seen in COVID-19 may contribute to NETosis, where neutrophils release extracellular traps, further driving inflammation and tissue damage [17].
AAT is the primary inhibitor of neutrophil elastase, a serine protease released by neutrophils, monocytes, macrophages, and mast cells during immune responses to kill pathogens [18]. Neutrophil elastase activates receptors to increase chemokines and proinflammatory cytokines, promoting leukocyte recruitment [19, 20]. Elevated elastase contributes to cell death, inflammation, pulmonary vascular permeability, and injury [21] and is linked to conditions like chronic obstructive pulmonary disease [22], acute lung injury [23], and acute respiratory distress syndrome [24]. The D614G mutation in the SARS-CoV-2 spike protein created a cleavage site for elastase, enhancing viral spread, particularly in AAT-deficient individuals [25].
Granzyme B, a serine protease in cytotoxic T and natural killer cells, shares structural similarities with chymotrypsin and may be inhibited by AAT [16]. These cells release granzyme B via exocytosis to induce rapid cell death in infected cells [26]. Granzyme B can also function extracellularly, promoting proinflammatory cytokine responses and tissue remodeling that can lead to tissue destruction [27–30]. As a pro-apoptotic factor, granzyme B activates cytokines like Interleukin-18, which amplifies the innate immune response [31].
To evaluate the therapeutic potential of granzyme B and neutrophil elastase, we assessed their anti-protease and complement activity in COVID-19 severity using functional mass spectrometry. Serpin inhibition involves a 1:1 complex formation that irreversibly inactivates both the serpin and protease [32, 33]. The RCL of the serpin targets specific proteases, mimicking their substrate sequence, leading to cleavage, a conformational change, and the permanent deactivation of both proteins [34].
Immunoassays cannot differentiate between active and inactivated serpins, but functional mass spectrometry can detect peptide sequences and assess the state of the RCL region [35]. This study identified distinct peptide sequences in the RCL regions of AAT and ACT, associated with elastase and granzyme B, respectively. Intact peptide sequences in the RCL represent active anti-protease activity. Protein depletion beads were used to remove albumin and IgG from serum, enriching lower-abundance proteins for proteomic analysis.
Our results show that AAT significantly increases within 48 h of hospital admission in COVID-19 patients, correlating with disease severity and elevated neutrophil elastase levels. This aligns with previous studies showing increased serpins, including AAT, in COVID-19 sera, particularly in patients with high Interleukin-6 levels [36], and evidence suggesting that AAT deficiency increases the risk of severe COVID-19 [37].
The anti-protease and anti-inflammatory activity of AAT [38] may help mitigate COVID-19 and the hyperactive immune response. AAT inhibits SARS-CoV-2 entry by inactivating TMPRSS2, a serine protease that primes the spike protein for cell surface fusion [39]. AAT also protects against cytotoxicity by inhibiting inflammatory proteases, particularly neutrophil elastase [36]. Our results show that within 48 h of hospital admission, ACT activity increased with COVID-19 infection, peaking in mild cases. In severe COVID-19, ACT activity decreased, likely due to higher granzyme B and other proteases, leading to greater proteolysis that may inactivate ACT and compromise its regulatory function.
ACT is a serpin that inhibits chymotrypsin-like proteases, including neutrophil cathepsin G and mast cell chymases [40, 41]. Its concentration increases during acute inflammation and may protect tissues, such as the lower respiratory tract and brain, from proteolytic damage [42]. In cell culture, ACT-cathepsin G complexes signal acute phase protein synthesis and increase interleukin-6 production by fibroblasts [43]. Since ACT irreversibly forms complexes with cathepsin G, further research is needed to determine whether ACT activity protects against or contributes to severe COVID-19.
ACT is also a substrate for neutrophil elastase and other proteases [44]. In these cases, the protease cleaves ACT without forming an irreversible complex, rendering ACT inactive. Cleaved ACT products become potent chemoattractants at nanomolar concentrations [45], with slower serum clearance than ACT-cathepsin G complexes, suggesting prolonged neutrophil chemotaxis and potential interleukin-6 (IL-6) stimulation. Elevated IL-6 is linked to acute COVID-19 symptoms, including complement cascade upregulation, and current COVID-19 treatment regimens include the use of the IL-6 specific monoclonal antibody Tocilizumab [36].
C3a, a small complement fragment, is an anaphylatoxin that regulates inflammation with both pro- and anti-inflammatory effects [46]. It triggers chemokine release through mast cell degranulation and increased vascular permeability, but also helps regulate B-cell immune responses. C3 cleavage to form C3a is a key step in all complement pathways, necessary for C3 convertase to generate the membrane attack complex (C5b-9), which induces cell lysis and death. At sublytic levels, C5b-9 regulates tissue homeostasis by influencing cell cycle, proliferation, and differentiation [47]. C3 can also be cleaved by various serine proteases, including thrombin and cathepsin, at sites of inflammation [48]. As an anaphylatoxin, C3a increases vascular permeability and causes vasodilation, which can result in hypotension. The latter requires vasoactive support.
Multiple studies suggest that complement activation plays a key role in COVID-19 pathology [49], with C3 split products (e.g., C3a) often monitored by immunoassay [50, 51]. Using functional mass spectrometry, we distinguished cleaved C3b sequences from unprocessed C3 and examined the C3a/C3 beta; ratio as a relative metric for C3b activation, with lower ratios indicating greater activation. A limitation of this method is the use of a small 9-amino acid C3a peptide (FISLGEACK) compared to the full 77-amino acid C3a. Future studies are needed to confirm whether this peptide is sufficient for monitoring C3 subforms and to compare results with immunoassay measurements.
Our findings align with previous studies, showing an inverse relationship between the C3a/C3 beta ratio and COVID-19 severity. This suggests that anti-complement therapy may benefit severe COVID-19 patients. The mass spectrometry approach allows for efficient examination of functional characteristics, such as post-translational modifications and genetic variations, which are challenging for immunoassays. By comparing anti-protease activity (AAT/ACT ratio) to complement activation (C3a/C3 beta ratio), mild COVID-19 cases clustered in the lower right quadrant, indicating low anti-protease activity and minimal complement activation. These clusters may provide insights into disease severity and help predict disease progression and optimal interventions.
The COVID-19 pandemic and long COVID highlight the need for therapeutics that target viral infection and modulate immune responses. Serine proteases and serpins are promising drug targets for immunomodulation [52]. Future studies on the balance between protease and anti-protease activities, particularly AAT/ACT ratios, may reveal insights into enzyme regulation, complement activation, and clinical outcomes. The AAT/ACT ratio could serve as both a biomarker for COVID-19 severity and a potential therapeutic target.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Western University, Human Research Ethics Board. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the ’ legal guardians/next of kin.
Author contributions
Conceptualization, DF and MK; methodology, DF, SR, MK, LV, GC, HZ, AS, and DR; formal analysis, DF and MK; resources, DF; original draft preparation, DF, MK, and MQ; review and editing, SR, LV, GC, HZ, AS, and DR; funding acquisition, DF. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. DF acknowledges funding from the London Health Sciences Foundation (https://lhsf.ca/) and the AMOSO Innovation Fund (INN20-029).
Acknowledgments
We thank the enthusiastic support of the frontline Nursing Staff and Research Assistants at London Health Sciences Centre.
Conflict of interest
Some authors (SR and MK) are employees of Biotech Support Group LLC, who provide proteomic and genomic sample prep and enrichment products and services.
The remaining authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Footnotes
References
1.
Diamond MS Kanneganti T-D . Innate immunity: the first line of defense against SARS-CoV-2. Nat Immunol (2022) 23(2):165–76. 10.1038/s41590-021-01091-0
2.
Peiris JS Chu C Cheng V Chan K Hung I Poon L et al Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. The Lancet (2003) 361(9371):1767–72. 10.1016/s0140-6736(03)13412-5
3.
Wong RSM . Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ (2003) 326(7403):1358–62. 10.1136/bmj.326.7403.1358
4.
Java A Apicelli AJ Liszewski MK Coler-Reilly A Atkinson JP Kim AH et al The complement system in COVID-19: friend and foe? JCI Insight (2020) 5(15):e140711. 10.1172/jci.insight.140711
5.
Fraser DD Cepinskas G Slessarev M Martin C Daley M Miller MR et al Inflammation profiling of critically ill coronavirus disease 2019 patients. Crit Care Explorations (2020) 2(6):e0144. 10.1097/cce.0000000000000144
6.
Akgun E Tuzuner MB Sahin B Kilercik M Kulah C Cakiroglu HN et al Proteins associated with neutrophil degranulation are upregulated in nasopharyngeal swabs from SARS-CoV-2 patients. PLOS ONE (2020) 15(10):e0240012. 10.1371/journal.pone.0240012
7.
Meyer-Hoffert U Wiedow O . Neutrophil serine proteases: mediators of innate immune responses. Curr Opin Hematol (2011) 18(1):19–24. 10.1097/moh.0b013e32834115d1
8.
Mantovani A Cassatella MA Costantini C Jaillon S . Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol (2011) 11(8):519–31. 10.1038/nri3024
9.
Medjeral-Thomas NR Troldborg A Hansen AG Pihl R Clarke CL Peters JE et al Protease inhibitor plasma concentrations associate with COVID-19 infection. Oxford Open Immunol (2021) 2(1):iqab014. 10.1093/oxfimm/iqab014
10.
Mansouri V Rezaiee Tavirani M Okhovatian F Allah Abbaszadeh H . Introducing markers which are involved in COVID-19 disease: severe condition versus mild state, a network analysis. J Cell and Mol Anesth (2022) 7(2):109–15. 10.22037/jcma.v6i4.35656
11.
Suvarna K Biswas D Pai MGJ Acharjee A Bankar R Palanivel V et al Proteomics and machine learning approaches reveal a set of prognostic markers for COVID-19 severity with drug repurposing potential. Front Physiol (2021) 12:652799. 10.3389/fphys.2021.652799
12.
Priestap F Kao R Martin CM . External validation of a prognostic model for intensive care unit mortality: a retrospective study using the ontario critical care information system. Can J Anesthesia/Journal canadien d'anesthésie (2020) 67(8):981–91. 10.1007/s12630-020-01686-5
13.
Singer M Deutschman CS Seymour CW Shankar-Hari M Annane D Bauer M et al The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA (2016) 315(8):801–10. 10.1001/jama.2016.0287
14.
Zahn-Zabal M Michel PA Gateau A Nikitin F Schaeffer M Audot E et al The neXtProt knowledgebase in 2020: data, tools and usability improvements. Nucleic Acids Res (2020) 48(D1):D328–D334. 10.1093/nar/gkz995
15.
Pino LK Searle BC Bollinger JG Nunn B MacLean B MacCoss MJ . The skyline ecosystem: informatics for quantitative mass spectrometry proteomics. Mass Spectrom Rev (2020) 39(3):229–44. 10.1002/mas.21540
16.
Wowk ME Trapani JA . Cytotoxic activity of the lymphocyte toxin granzyme B. Microbes Infect (2004) 6(8):752–8. 10.1016/j.micinf.2004.03.008
17.
Al-Kuraishy HM Al-Gareeb AI Al-hussaniy HA Al-Harcan NAH Alexiou A Batiha GES . Neutrophil extracellular traps (NETs) and Covid-19: a new frontiers for therapeutic modality. Int Immunopharmacology (2022) 104:108516. 10.1016/j.intimp.2021.108516
18.
Korkmaz B Horwitz MS Jenne DE Gauthier F Sibley D . Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol Rev (2010) 62(4):726–59. 10.1124/pr.110.002733
19.
Pham CT . Neutrophil serine proteases: specific regulators of inflammation. Nat Rev Immunol (2006) 6(7):541–50. 10.1038/nri1841
20.
Pham CT . Neutrophil serine proteases fine-tune the inflammatory response. The Int J Biochem & Cell Biol (2008) 40(6-7):1317–33. 10.1016/j.biocel.2007.11.008
21.
Voisin MB Leoni G Woodfin A Loumagne L Patel NS Di Paola R et al Neutrophil elastase plays a non‐redundant role in remodeling the venular basement membrane and neutrophil diapedesis post‐ischemia/reperfusion injury. The J Pathol (2019) 248(1):88–102. 10.1002/path.5234
22.
Owen C . Roles for proteinases in the pathogenesis of chronic obstructive pulmonary disease. Int J Chronic Obstructive Pulm Dis (2008) 3:253–68. 10.2147/copd.s2089
23.
Lee WL Downey GP . Leukocyte elastase: physiological functions and role in acute lung injury. Am J Respir Crit Care Med (2001) 164(5):896–904. 10.1164/ajrccm.164.5.2103040
24.
Moraes TJ Chow CW Downey GP . Proteases and lung injury. Crit Care Med (2003) 31(4 Suppl. l):S189–94. 10.1097/01.ccm.0000057842.90746.1e
25.
Bhattacharyya C Das C Ghosh A Singh AK Mukherjee S Majumder PP et al SARS-CoV-2 mutation 614G creates an elastase cleavage site enhancing its spread in high AAT-deficient regions. Infect Genet Evol (2021) 90:104760. 10.1016/j.meegid.2021.104760
26.
Cullen SP Martin SJ . Mechanisms of granule-dependent killing. Cell Death & Differ (2008) 15(2):251–62. 10.1038/sj.cdd.4402244
27.
Buzza MS Zamurs L Sun J Bird CH Smith AI Trapani JA et al Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J Biol Chem (2005) 280(25):23549–58. 10.1074/jbc.m412001200
28.
Isaaz S Baetz K Olsen K Podack E Griffiths GM . Serial killing by cytotoxic T lymphocytes: T cell receptor triggers degranulation, re-filling of the lytic granules and secretion of lytic proteins via a non-granule pathway. Eur J Immunol (1995) 25(4):1071–9. 10.1002/eji.1830250432
29.
Spaeny-Dekking EH Hanna WL Wolbink AM Wever PC Kummer AJ Swaak AJG et al Extracellular granzymes A and B in humans: detection of native species during CTL responses in vitro and in vivo. The J Immunol (1998) 160(7):3610–6. 10.4049/jimmunol.160.7.3610
30.
Wensink AC Hack CE Bovenschen N . Granzymes regulate proinflammatory cytokine responses. The J Immunol (2015) 194(2):491–7. 10.4049/jimmunol.1401214
31.
Akeda T Yamanaka K Tsuda K Omoto Y Gabazza EC Mizutani H . CD8+ T cell granzyme B activates keratinocyte endogenous IL-18. Arch Dermatol Res (2014) 306(2):125–30. 10.1007/s00403-013-1382-1
32.
Huntington JA Carrell RW . The serpins: nature's molecular mousetraps. Sci Prog (2001) 84(Pt 2):125–36. 10.3184/003685001783239032
33.
Khan MS Singh P Azhar A Naseem A Rashid Q Kabir MA et al Serpin inhibition mechanism: a delicate balance between native metastable state and polymerization. J Amino Acids (2011) 2011:1–10. 10.4061/2011/606797
34.
Marijanovic EM Fodor J Riley BT Porebski BT Costa MGS Kass I et al Reactive centre loop dynamics and serpin specificity. Scientific Rep (2019) 9(1):3870. 10.1038/s41598-019-40432-w
35.
Tsutsui Y Sarkar A Wintrode PL . Probing serpin conformational change using mass spectrometry and related methods. Methods Enzymol (2011) 501:325–50. 10.1016/b978-0-12-385950-1.00015-8
36.
D’Alessandro A Thomas T Dzieciatkowska M Hill RC Francis RO Hudson KE et al Serum proteomics in COVID-19 patients: altered coagulation and complement status as a function of IL-6 level. J Proteome Res (2020) 19(11):4417–27. 10.1021/acs.jproteome.0c00365
37.
Bai X Schountz T Buckle AM Talbert JL Sandhaus RA Chan ED . Alpha-1-antitrypsin antagonizes COVID-19: a review of the epidemiology, molecular mechanisms, and clinical evidence. Biochem Soc Trans (2023) 51(3):1361–75. 10.1042/bst20230078
38.
Brantly ML Paul LD Miller BH Falk RT Wu M Crystal RG . Clinical features and history of the destructive lung disease associated with alpha-1-antitrypsin deficiency of adults with pulmonary symptoms. Am Rev Respir Dis (1988) 138(2):327–36. 10.1164/ajrccm/138.2.327
39.
Wettstein L Weil T Conzelmann C Müller JA Groß R Hirschenberger M et al Alpha-1 antitrypsin inhibits TMPRSS2 protease activity and SARS-CoV-2 infection. Nat Commun (2021) 12(1):1726. 10.1038/s41467-021-21972-0
40.
Korkmaz B Attucci S Jourdan ML Juliano L Gauthier F . Inhibition of neutrophil elastase by α1-protease inhibitor at the surface of human polymorphonuclear neutrophils. The J Immunol (2005) 175(5):3329–38. 10.4049/jimmunol.175.5.3329
41.
Kalsheker NA . α1-antichymotrypsin. The Int J Biochem & Cell Biol (1996) 28(9):961–4. 10.1016/1357-2725(96)00032-5
42.
Calvin J Price CP . Measurement of serum α1-antichymotrypsin by immunoturbidimetry. Ann Clin Biochem (1986) 23(Pt 2):206–9. 10.1177/000456328602300212
43.
Kurdowska A Travis J . Acute phase protein stimulation by alpha 1-antichymotrypsin-cathepsin G complexes. Evidence for the involvement of interleukin-6. J Biol Chem (1990) 265(34):21023–6. 10.1016/s0021-9258(17)45321-x
44.
Rubin H Plotnick M Wang Z Liu X Schechter NM Zhong Q et al Conversion of.alpha.1-Antichymotrypsin into a human neutrophil elastase inhibitor: demonstration of variants with different association rate constants, stoichiometries of inhibition, and complex stabilities. Biochemistry (1994) 33(24):7627–33. 10.1021/bi00190a016
45.
Potempa J Fedak D Dubin A Mast A Travis J . Proteolytic inactivation of alpha-1-anti-chymotrypsin. Sites of cleavage and generation of chemotactic activity. J Biol Chem (1991) 266(32):21482–7. 10.1016/s0021-9258(18)54664-0
46.
Coulthard LG Woodruff TM . Is the complement activation product C3a a proinflammatory molecule? Re-evaluating the evidence and the myth. The J Immunol (2015) 194(8):3542–8. 10.4049/jimmunol.1403068
47.
Tegla CA Cudrici C Patel S Trippe R III Rus V Niculescu F et al Membrane attack by complement: the assembly and biology of terminal complement complexes. Immunol Res (2011) 51(1):45–60. 10.1007/s12026-011-8239-5
48.
Markiewski MM Nilsson B Nilsson Ekdahl K Mollnes TE Lambris JD . Complement and coagulation: strangers or partners in crime?Trends Immunol (2007) 28(4):184–92. 10.1016/j.it.2007.02.006
49.
Zelek WM Harrison RA . Complement and COVID-19: three years on, what we know, what we don't know, and what we ought to know. Immunobiology (2023) 228(3):152393. 10.1016/j.imbio.2023.152393
50.
Detsika MG Diamanti E Ampelakiotou K Jahaj E Tsipilis S Athanasiou N et al C3a and C5b-9 differentially predict COVID-19 progression and outcome. Life (Basel) (2022) 12(9):1335. 10.3390/life12091335
51.
Ma L Sahu SK Cano M Kuppuswamy V Bajwa J McPhatter J et al Increased complement activation is a distinctive feature of severe SARS-CoV-2 infection. Sci Immunol (2021) 6(59):eabh2259. 10.1126/sciimmunol.abh2259
52.
Lucas A Liu L Dai E Bot I Viswanathan K Munuswamy-Ramunujam G et al The serpin saga; development of a new class of virus derived anti-inflammatory protein immunotherapeutics. Adv Exp Med Biol (2009) 666:132–56. 10.1007/978-1-4419-1601-3_11
Summary
Keywords
COVID-19, functional mass spectrometry, neutrophil elastase, lymphocyte granzyme B, complement
Citation
Fraser DD, Roy S, Kuruc M, Quintero M, Van Nynatten LR, Cepinskas G, Zheng H, Soherwardy A and Roy D (2025) Functional mass spectrometry indicates anti-protease and complement activity increase with COVID-19 severity. Exp. Biol. Med. 250:10308. doi: 10.3389/ebm.2025.10308
Received
06 July 2024
Accepted
14 January 2025
Published
29 January 2025
Volume
250 - 2025
Updates
Copyright
© 2025 Fraser, Roy, Kuruc, Quintero, Van Nynatten, Cepinskas, Zheng, Soherwardy and Roy.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Douglas D. Fraser, douglas.fraser@lhsc.on.ca
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.