Abstract
Messenger RNA (mRNA) therapeutics have significantly transformed contemporary medicine, particularly through their role as the active component in the SARS-CoV-2 vaccine. This remarkable achievement is the culmination of extensive research conducted over many years by scientists. The widespread administration of the COVID-19 vaccine has further accelerated research into the precise therapeutic potential of mRNA technologies. Since mRNA doesn’t integrate with the host genome, the safety and versatility of mRNA-based therapeutics make them an iconic candidate in targeted therapies. Due to a surge in innovation efforts, biomodification of the molecular signatures of mRNAs like the 5′cap, untranslated regions (UTRs), and the poly(A) tail are being developed to increase translation efficacy. Recent advancements in chemical modifications, codon optimization techniques, and targeted delivery methods have significantly enhanced the stability of synthetic mRNAs while concurrently reducing their immunogenicity. Various mRNA manufacturing and synthesizing methods are investigated in this review, focusing on their scalability and limitations. mRNA therapeutic strategies can be divided into protein replacement, immune modulation, and cellular modulation. This review explores mRNA’s molecular landscape and comprehensive utility, including applications in both clinical trials and commercial sectors.
Impact statement
The development of messenger RNA (mRNA) therapeutics is currently of paramount importance. Its successful application in SARS-CoV-2 vaccines has catalysed rapid advancements in research, demonstrating both its safety and versatility. Ongoing innovations in mRNA design continue to enhance its therapeutic potential. Beyond vaccines, mRNA is revolutionising medicine through a variety of applications, including protein replacement therapy, advanced cancer treatments such as personalised vaccines, and therapies for other infectious diseases. Additionally, it shows promise for addressing genetic and metabolic disorders. This broad and evolving adoption of mRNA display the significance and ongoing impact of mRNA technology on global health. The development of mRNA therapeutics is currently of paramount importance. Its successful application in SARS-CoV-2 vaccines has catalyzed rapid advancements in research, demonstrating both its safety and versatility. Ongoing innovations in mRNA design continue to enhance its therapeutic potential. Beyond vaccines, mRNA is revolutionizing medicine through a variety of applications, including protein replacement therapy, advanced cancer treatments such as personalized vaccines, and therapies for other infectious diseases. Additionally, it shows promise for addressing genetic and metabolic disorders. This broad and evolving adoption of mRNA displays the significance and ongoing impact of mRNA technology on global health. The article is directed towards the broader scientific community, particularly early to mid-career researchers and professionals seeking to understand therapeutic development and the commercial landscape of mRNA technology. By providing a comprehensive overview of drug design, delivery systems, commercial applications, and patent insights, the document serves as a complete resource for mRNA-related information. This makes it valuable for individuals aiming to understand the current state, future directions, and applications within this rapidly advancing field. Several review articles have examined mRNA therapeutics, particularly due to its recent popularity within the scientific community. Among these, a few stand out, notably Quin et al., “mRNA-based therapeutics: powerful and versatile tools to combat diseases,” and Lu et al., “Current landscape of mRNA technologies and delivery systems for new modality therapeutics.” The proposed review article, titled “Beginning of a New Era of Synthetic Messenger RNA (mRNA) Therapeutics: Comprehensive Insights on mRNA Drug Design, Development, and Applications,” is uniquely distinguished from the aforementioned reviews due to its explicit focus on current therapeutic and commercial applications, analyzed through published patents and clinical trial records. While all three articles provide comprehensive reviews of mRNA development, the proposed article is notable for its specific exploration of mRNA’s utility in commercial sectors and its direct incorporation of insights derived from published patents. This approach offers a distinct understanding of the intellectual property and market landscape of mRNA technologies, which is not explicitly detailed in the other two reviews. Furthermore, the proposed review article delves deeply into understanding the versatility of mRNA therapeutics, particularly in their therapeutic application to address the molecular pathogenesis of conditions underlying the aforementioned commercial inventions.
Introduction
Nucleic acid therapeutics are therapies formulated by deoxyribonucleic acids (DNAs) and/or ribonucleic acids (RNAs) as their key bioactive compound, inducing its therapeutic effect by modulating gene expression [1]. During recent decades, DNA and RNA-based therapies have gained significant attention as viable therapeutic options to treat a variety of health conditions including infectious diseases, cancer, neurodegeneration, metabolic disorders, and rare genetic diseases [2, 3]. Following the discovery of mRNA in 1961, scientists began to investigate its chemical properties and potential therapeutic applications (Figure 1). RNAs perform diverse yet precise functions in cellular metabolism within the human body. Approximately 75% of human genome is transcribed into RNAs [4]. RNAs can be broadly classified into 3 categories: Parasitic (self-propagating retrotransposons, viral RNA and CRISPR RNA), Regulatory and Protein-coding associated RNAs [5]. Three protein coding associated RNAs that plays a key role in protein synthesis are messenger RNA (mRNA), Transfer RNA (tRNA) and Ribosomal RNA (rRNA). mRNA plays a critical role in gene expression [4, 5]. Compared to DNA-based vaccines, mRNA therapy offers several advantages. The mRNAs’ transient nature and ability to be temporally controlled, while not integrating into genome which reduces the risk of carcinogenesis and mutagenesis, mRNA is potentially a safer alternate to traditional gene therapy. And also, mRNA only requires to be delivered into the cytoplasm of target cells without the need for direct delivery into the nucleus to initiate action, making mRNA a promising/logical therapeutic candidate [1, 4, 6, 7]. Wolff et al. introduced exogenous RNA and DNA to cells to investigate the increase in expression [8]. Later in 1993, a study conducted by Martinon et al. was the first time mRNA was used to introduce virus-specific cytotoxic T lymphocytes in animals [9]. This study paved the way for the mRNA vaccines that we currently use today [9]. The COVID-19 pandemic triggered a major shift in the use of mRNA for therapeutics, accelerating both research and public awareness [3].
FIGURE 1
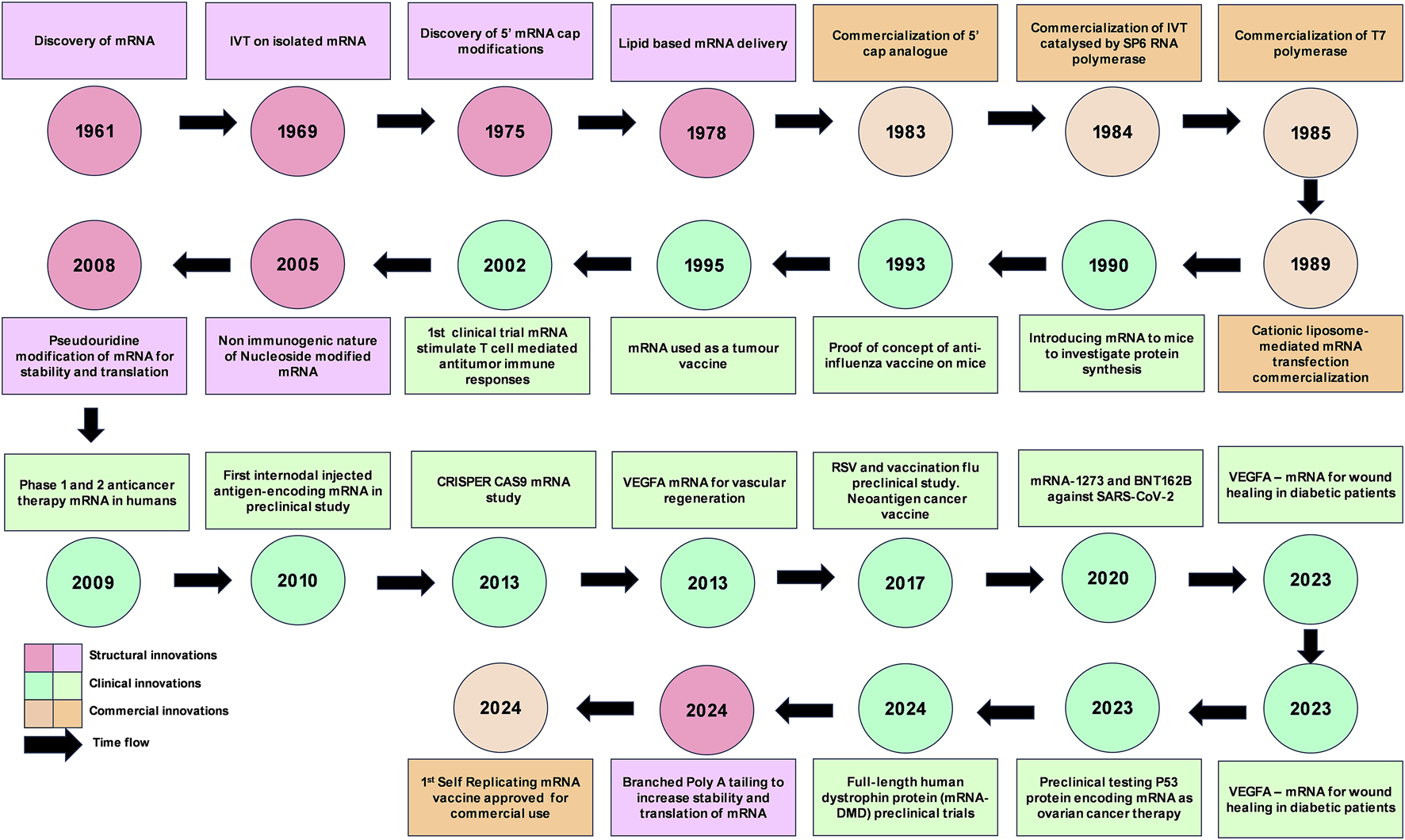
mRNA innovations timeline. IVT, in vitro transcription; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; RSV, respiratory syncytial virus; CRISPR, clustered regularly interspaced short palindromic repeats; CRISPR-Cas9, CRISPR-associated protein 9; VEGFA, Vascular endothelial growth factor A; DMD, Duchenne muscular dystrophy.
Manufacturing exogenous mRNA has several benefits, primarily being more convenient, cost-effective, and final end-products not being subjected to additional regulations and scrutiny associated with genetically modified organisms [10]. However, mRNA treatments have drawbacks including delivery, stability and documented immunological side effects [11]. Exogenous mRNA is known to be cleaved by RNase, resulting in activation of Toll-like receptors (TLRs) 3, 7 and 8 inducing innate immune response that causes cytokine-mediated toxicity [6]. However, the possibility of reducing the side effects of immunogenic activity was demonstrated by Weissman and Kariko, in which modified mRNA (modRNA) was synthesized by incorporating modified ribonucleotides to reduce immunogenic activity while increasing the level of protein expression [12–14].
In fact, one of the most well-known mRNA vaccines against SARS-CoV-2 has a current count of more than 13.72 billion doses administered worldwide [15]. The present review will highlight the noteworthy uses of mRNA within the context of therapeutic and commercial applications in recent times (post 2019). Prior to this, mRNA structure, mRNA design, mRNA synthesis, molecular mechanism and therapeutic strategies will be explored.
mRNA design and modifications
mRNA is a polymer of ribonucleotide building blocks, each composed of a ribose sugar, a phosphate group, and a nitrogenous base (adenine, guanine, uracil, or cytosine) [16]. Based on the principle of central dogma of life, the fundamental instructions of protein synthesis contained in DNA is expressed by transcribing the information onto mRNA. In prokaryotes, mRNA is directly transcribed from DNA and translated using ribosomes identifying the Shine-Dalgarno sequence and assembling across it in 5′UTR of the mRNA. In contrast, eukaryotes use RNA polymerase II to synthesize precursor mRNA (pre-mRNA), which is then modified to achieve the final mature mRNA. The mature mRNA is then transported to cytoplasm and translated into protein [17]. An mRNA consists of five main structural elements, namely, 5′cap, 5′untranslated region (UTR), protein coding sequence, 3′UTR, and poly adenine (A) tail (Figure 2) [10].
FIGURE 2
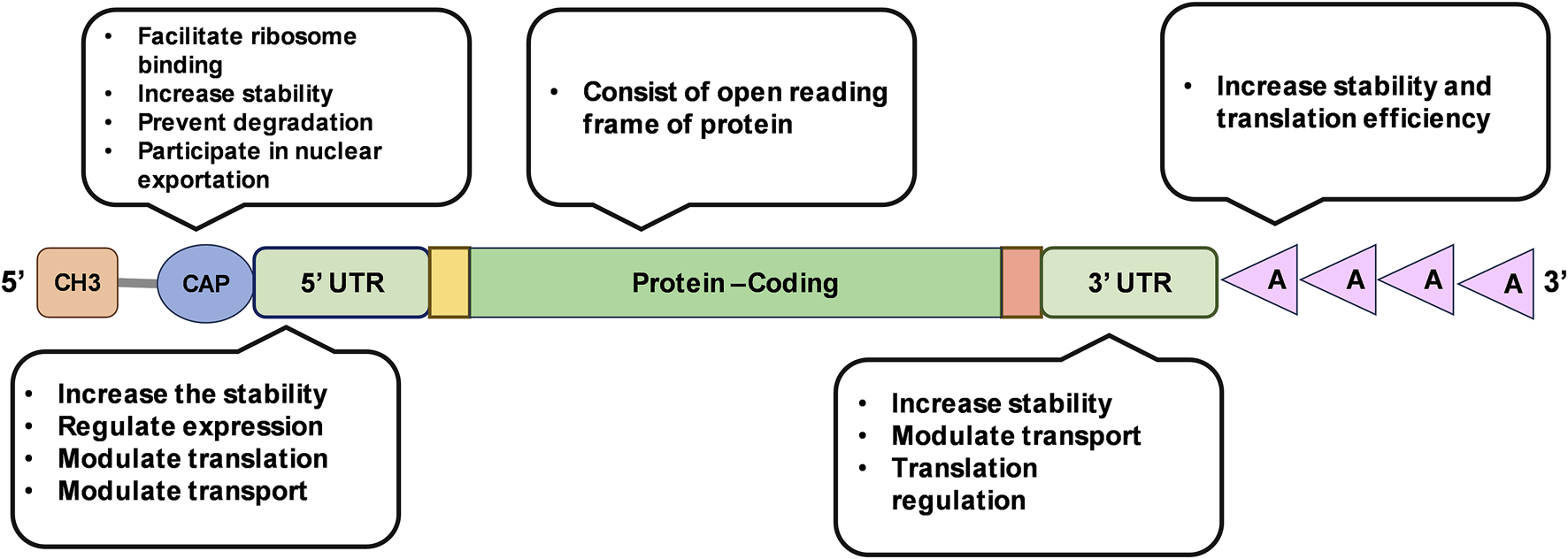
Illustration of mRNA structure and its functional features.
Nucleotide modification
Nucleotides govern the dynamics of the expression of the genetic information encoded in the mRNA and they can be modified enzymatically or non-enzymatical (alkylation, oxidation or UV damage), to alter the biological properties of mRNA. In therapeutic and commercial applications of exogenous mRNA these modifications (Figure 3) are made in order to increase translation initiation, improve expression and reduce immunogenicity [14, 16, 18].
FIGURE 3
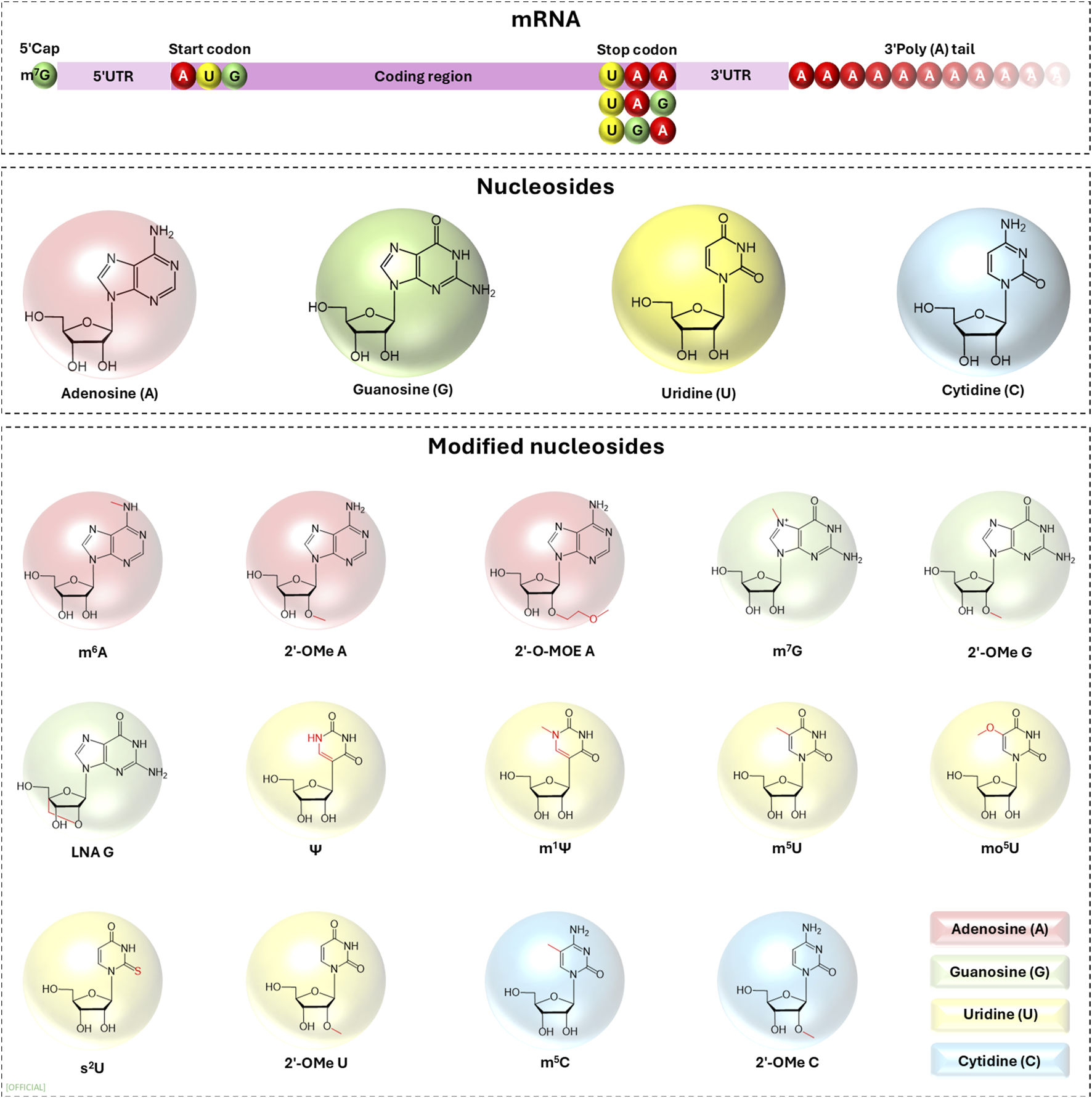
Examples of nucleoside modifications that have been investigated for constructing in vitro transcribed mRNA. UTR, untranslated region; m6A, N6-methyladenosine; m7G, N7-methylguanosine; Ψ, pseudouridine; m1Ψ, N1-methylpseudouridine; m5U, 5-methyluridine; mo5U, 5-methoxyuridine; s2U, 2-thiouridine; m5C, 5-methylcytosine; 2′-O-methyl, 2′-OMe; 2′-O-MOE, 2′-O-methoxyethyl; LNA, locked nucleic acid.
It is recorded that innate immune system differentiates self-originating mRNA from IVT mRNA triggering immune responses via pattern recognition receptors (PRRs). The PRRs that activate immune responses are Toll-like receptors (TLRs) and retinoic acid inducible gene I (RIG-I) like receptors (RLRs). This activation of the immune responses against IVT mRNA severely affect the translation as well as induce severe immunological side effects to the patient making IVT mRNA an unviable therapeutic pursuit. This was mitigated by the breakthrough discovery made by Weissman and Kariko who were awarded the Nobel prize in 2023 [19]. Weissman and Kariko explored the possibility of using modified nucleotides (m5C, m6A, m5U, s2U or pseudouridine) in IVT mRNA to reduce immunogenicity of IVT mRNA. They discovered that using modified nucleotides (m5C, m6A, m5U, s2U or pseudouridine) in IVT mRNA, ablates the activation of TLR3, TLR7, TLR8 and TLR9 leading to a significant reduction in the production of type I interferons (IFNs) and pro-inflammatory cytokines compared to unmodified IVT mRNA (Figure 3) [20, 21]. According to Weissman and Kariko, translation of Ψ-containing transcripts was 4-5-fold greater than that of unmodified transcripts in wild-type mouse embryonic fibroblasts (MEFs), yet in PKR knockout (PKR−/−) MEFs, the extent of translation of Ψ-modified mRNA was not different from that of unmodified mRNA. Thus, displaying that Ψ-mRNA translational increase is dependent on PKR activity [6, 14]. In the following year they identified that IVT mRNA containing uridine has a higher tendency to activate RNA-dependent protein kinases (PKR) which then phosphorylates translation initiation factor 2-alpha, inhibiting translation. The activation of PKR is limited in IVT mRNA containing pseudouridine (ψ) modification, thus increasing the translation relative to unmodified IVT mRNA [14]. During in vivo studies, 8-week-old BALB/c mice (weighing 18–23 g) was treated with 3.0 µg of Ψ-modified mRNA and unmodified mRNA it was observed that, unmodified mRNA induced high levels of IFN-α in serum while Ψ-modified mRNA did not induce higher level of IFN-α, 12-fold higher protein expression was observed [6].
Following the Kariko paradigm, Andries et al. identified that N1-methylpseudouridine (m1Ψ) alone or in combination with 5-methylcytidine (m5C) modification, displayed comparatively higher translation rates and increased stability. The modifications have shown a 13-fold increase in expression in single modified mRNA and 44-fold increase in double modified mRNA expression in mammalian cells and mice. They observed that (m5C/)m1Ψ-modified mRNA triggered a reduced innate immune response compared to (m5C/)Ψ-modified mRNA post-transfection making it a new benchmark for mRNA modification [22]. Following these inventions, in 2020 Pfizer-BioNTech m1Ψ-modification was used in SARS-CoV-2 mRNA vaccine [23]. Like Ψ-uridine, m1Ψ-uridine also evades processing by RNase T2, PLD3, and PLD4. But N1- m1Ψ-uridine is capable of directly activating TLR8. This selective activation activating TLR8 pathway without broader immune activation is leveraged in vaccine development and other immunotherapeutic strategies [24].
However, Mulroney et al. discovered that N1 methyl Ψ-modification (m1Ψ) causes +1 ribosomal frameshifting during IVT mRNA translation leading to the unintended creation of altered proteins with off-target immune responses. Even though there is no noted adverse effects arising from mistranslation, this exhibits room for improvement in mRNA design optimization [25].
Codon and sequence optimization
Codons are three-letter nucleotide sequences that correspond to amino acids and stop signals during protein synthesis. There are 64 codons that are specific to 20 amino acid building blocks and three stop codons. This nature of degeneracy is a feature of genetic code in all organisms. Apart from degeneracy, other key features of codons are being non-overlapping, non-ambiguous and universal among all living organisms [26]. The nature of degeneracy of genetic code creates the existence of synonymous codons that encode the same amino acid [27]. The rate of decoding these synonymous codons are biased, which is evident by the disproportionate representation in certain codons over other types in the transcriptome [28]. The codons with less than 1% frequency in the genome is known as a rare codon. It is observed that the cognate tRNA associated with the rare codons are less abundant in organisms and are referred to as rare tRNAs [29]. The efficiency of the selection of cognate tRNA from the cytoplasmic tRNA pool determines whether it is optimal or not. Hence, codon optimality is governed by the tRNA availability and the stochastic kinetics of the ribosomes [28]. A genome-wide analysis of mRNA decay rates conducted by Presnyak et al. revealed that mRNAs composed predominantly of optimal codons are on average more stable than the mRNAs containing predominantly non-optimal codons [30]. On the contrary, Saikia et al. study identifies that rare codons promote selective protein synthesis rather than affecting mRNA stability in mammalian systems during amino acid (aa) starvation. They identified that genes enriched in non-optimal codons [like ubiquitin-proteasome system (UPP)] maintained or even up-regulate their translation efficiency while ribosomal protein genes which are rich in common codons, experience translational suppression during aa starvation. The underlying mechanism involves the selective charging of rare tRNA isoacceptors which maintains translation, while abundant tRNA isoacceptors are depleting during starvation. Experimental validation was conducted using FLuc (firefly luciferase) reporters, with rare codons in UPP gene sequences enable sustained translation during amino acid starvation. Yet when rare codons were changed to common codons, the protein levels have been reduced. The team further discovered that the above translational effects had occurred while total mRNA levels were unchanged, indicating that codon optimality regulates protein synthesis at the translational level rather than through mRNA degradation or stabilization [31].
In molecular therapeutics, codon optimization is used to increase the expression and stability of recombinant proteins by avoiding the rare codons which can restrict protein synthesis. It has been identified that species with large population sizes and highly expressed genes are most likely to be encoded with codons with abundant cognate tRNA. And usage of codons with more abundant cognate tRNA significantly increases the protein expression more than 1000-fold [27]. However, this correlation is not well-established in humans. As study conducted by Parmley and Hyynen (2009) shows that humans have clusters of codons with low abundance cognate tRNA (rare cognate tRNA), hinting at novel mechanism of tissue-specific transcription regulation [27, 32]. It has been also identified that affecting codon optimality by having non-optimal codons is directly proportional to the instability of the mRNA [28]. Following the above revelations, Wang et al. engineered rare codon devices to manipulate the expression levels of proteins in E. coli. In their study, codon devices were utilized to manipulate four genes in fatty acid synthesis pathway, where they successfully increased the fatty acid yield by approximately two-fold [33].
The COVID-19 pandemic has accelerated the research in codon optimization where scientists are now exploring a variety of methods to design more stable mRNAs. Zhang et al. used classical lattice parsing in computational linguistic to develop an in silico algorithm of Linear Design to optimize structural stability and codon usage in mRNAs. Using the Linear Design algorithm, they developed novel designs for SARS-CoV-2 spike protein and varicella-zoster virus glycoprotein E protein. Relative to benchmarks, these novel mRNA designs have exhibited 2.3 - 2.9 fold increase in protein expression and a 128-fold increase in antibody response for vaccines, demonstrating the importance of UTR engineering and codon optimization [34]. Diez et al., has developed the algorithm known as iCodon which is a machine learning model trained with mRNA stability from zebrafish, Xenopus embryos, human cell lines, and mouse embryonic stem cells, to predict mRNA stability based on codon composition. During the study the team has restored the function of optimized variants of the slc45a2 gene, injected into albino zebrafish embryos, successfully restored pigmentation. Diez and the team has also improved the fluorescence intensities of AausFP1 protein in human 293T cells. iCodon-optimized variant had displayed 4-fold increase in fluorescence intensity compared to the original AausFP1 [35].
Research has displayed that maximal codon optimization doesn’t always yield the best translational results, over- optimization could lead to synthesis of functionally and structurally altered proteins [27]. Therefore, the current consensus among scientist is to approach codon optimization holistically taking into consideration of evolutionary pressure and natural selection [36].
5′Cap
The 5′end of mRNA possesses a unique structure known as a cap, which is composed of a 7-methylguanosine (m7G) linked to the mRNA via 5′-5′ triphosphate bridge (ppp). This particular cap is identified as “cap 0.” The 5′cap plays a fundamental role in protecting eukaryotic mRNA from premature exonuclease cleavage (Dcp 1/2) and assists mRNA transport and cap-dependent translation [37]. In the pre-mRNA, the 5′cap facilitates polyadenylation, nuclear export, and splicing by recruiting specific protein factors [38]. In the mature mRNA, it recruits eukaryotic translation initiation factor 4E (elF4E) via hydrophobic cation–Π interactions and electrostatic negative interactions of the triphosphate bridge. The cap is essential for the self- and non-self-discrimination of cellular RNAs to initiate innate immune responses against foreign RNAs. The mechanics of innate immune activation is by retinoic acid-inducible gene I receptor (RIG-I). The immunogenicity of cap zero is reduced by 2′-O methylation of the ribose sugar of the first (R1) or second (R2) nucleotide (Figure 4), creating cap 1 and cap 2 [39]. Cap 1 structure characterized by and often 2′-O-methylated, 7-methylguanosine linked to the first transcribed nucleotide [40, 41]. Despic and Jaffrey (2023) discovered that C1 is modified to C2 in the cytoplasm by cap methyltransferase 2 (CMTR2) in mRNA with a longer half-life [42]. The dual methylation in cap 2 enhances the stability and protein expression in transcripts by reducing the binding ability of the cap one mRNA to RIG-I [42].
FIGURE 4
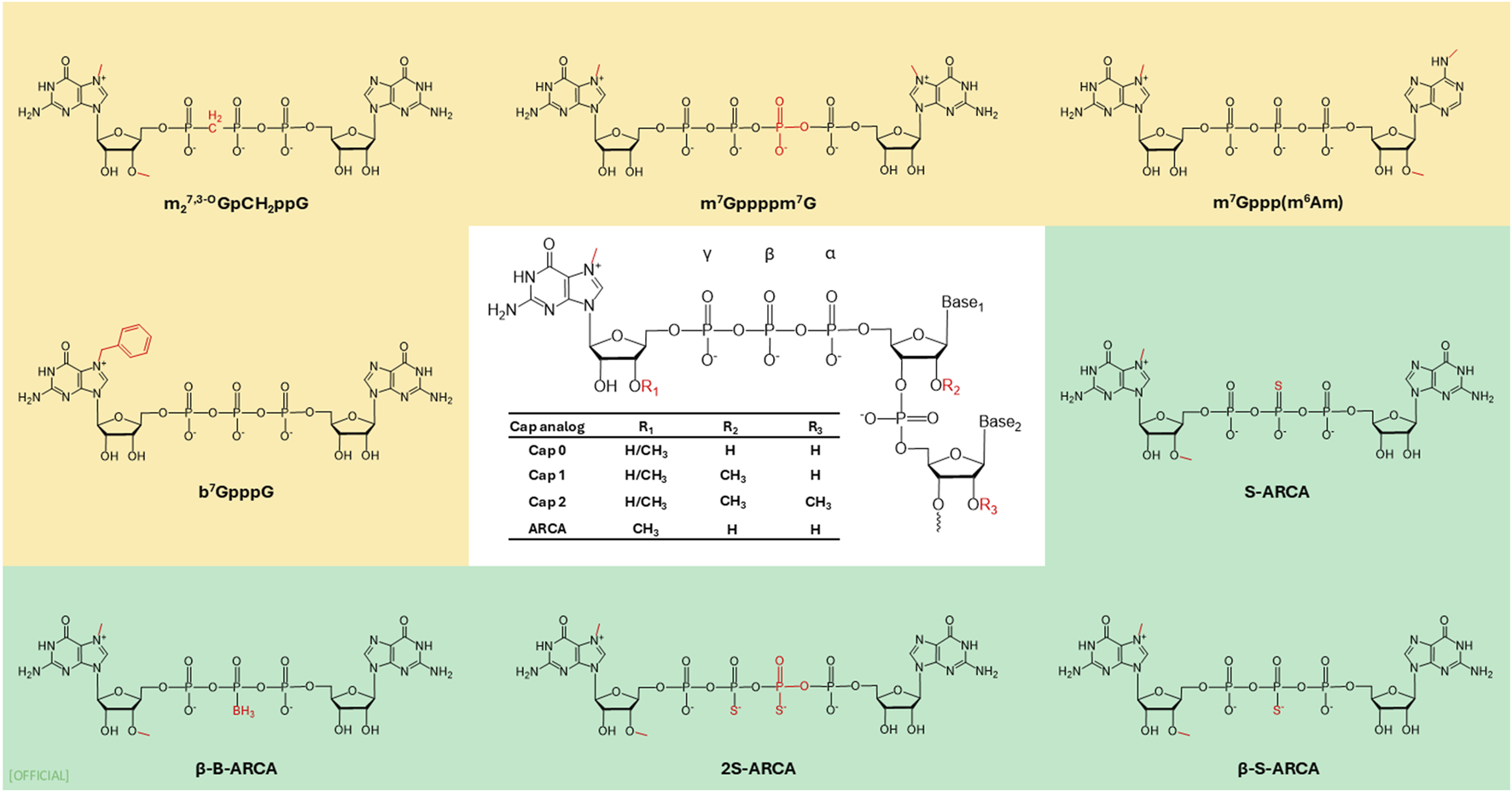
Examples of 5′cap modifications. ARCA: anti-reverse cap analog.
In cells, 5′Cap is introduced to mRNA by three sequential enzyme reactions [43]. In vitro transcribed (IVT) mRNA undergoes a capping reaction, which can be executed either post-transcriptionally or co-transcriptionally. Post-transcriptional capping is commonly done by a recombinant viral capping enzyme, such as vaccina virus-capping enzyme (VCE). VCE catalyses the capping reaction where standard eukaryotic m7GpppG structure is added to mRNA. Co-transcriptional capping is a reaction where a synthetic cap is added during transcription. This method provides versatility of using different cap structures yet has the drawback of potential synthesis of partially capped mRNA. Partial capping could take place due to, competition with GTP in IVT as well as cap binding protein hindered by incorrect orientation of the cap. To mitigate this, the anti-reverse cap analogue (ARCA: 3′-O-Me-m7G(5′)ppp(5′)G) was developed [19, 44]. Compared to ARCA which is a Cap 0 structure, CleanCap® on the other hand is co-transcriptional cap with cap 1 structure has mitigated with drawbacks of Cap 0: ARCA Cap, while being 94% capping efficiency [45]. A noteworthy method in increasing the rate of translation involves replacing the m7G with 7-benzylated guanosine. It has been reported that attaching the m7G with another m7G via tetraphosphate (m7Gppppm7G) increases the affinity of the mRNA with elF4E compared to the natural analogue, thereby increasing the translation by two fold [104]. Furthermore, Grezela et al. have modified this base cap by N2 modification to further increase translation and stability [46]. This concept of synthetically modified caps is seen in FDA-approved SARS-CoV-2 vaccines, with a cap analogue 7mG[5']ppp[5']NlmpNp increasing translation rates [104]. Pfizer-BioNTech and Moderna utilize cap 1 analogue in their vaccines [47]. However, Pfizer-BioNTech uses synthetic chemical process, while Moderna’s Cap 1 analogue is obtained through a chemical reaction catalysed by Vaccinia capping enzyme and Vaccinia 2′-O-methyltransferase [47, 48]. Warminski et al., (2024) have demonstrated that combination of trinucleotide capping with modifications in phosphate groups in cap structure is a potential avenue to develop novel high quality synthetic mRNA cap structures [43]. To further improve translation efficiency Senthilvelan et al., synthesised a novel locked nucleic acid (LNA)-modified trinucleotide cap analogue m7(LNA)G(5′)ppp(5′)AmpG, which has increased the translation efficiency of mRNA in vitro by factor of 5 relative to the ARCA-capped mRNA. By LNA modification and the C3′-endo (N-type) conformation in the LNA structure has increased intracellular stability, resulting in increased translation efficiency [49]. Another promising development is photocaged 5′cap analogues in the context of cap structures. This specific cap analogue serves to regulate expression by spatiotemporal control, thus further refining the targeting and activation of genes. Klöcker et al., have developed this 5′ Cap with photo-cleavable groups called “Flash Caps” which inhibit translation by blocking the elF4E binding. During in vitro studies when exposed to laser irradiation cleaving the Flash Cap group restores Cap 0 translation. This development creates a novel approach to regulate expression by spatiotemporal control [50]. Tarbashevich et al., has displayed light-controlled protein expression in Zebrafish embryo in vivo model, where they were able to induce the expression of fluorescent proteins for cell labelling, toxins for targeted cell ablation, and morphogens like Bmp2b and β-catenin to influence embryonic development [51]. The advancement in molecular structures of 5′cap not only enables production of more stable mRNA with higher expression levels, but it has also created new avenues to regulate gene expression.
The untranslated region (UTR)
UTR is non-coding sequences located at 5′and 3′of the coding regions of the mature mRNA. Current knowledge stipulates that UTRs regulate localization, translation, and stability of mRNAs. It has been identified that the length, start-site consensus sequence, secondary structures, upstream open reading frame, upstream the canonical AUG, internal ribosome entry site (IRES) and sequences that bind regulatory proteins present in 5′UTR are responsible for the regulation of mRNA translation rate [52]. The 5′UTR plays a crucial role in initiation of translation by recruitment of the ribosome and initiation factors to assemble the pre-initiation complex. The 5′cap identifies and recruits elF4A initiation factors essential for the translation. In addition, 5′UTR in some instances contains internal ribosome entry sites which recruits ribosome and initiate cap-independent translation. The 5′UTR also plays a crucial role in the regulation of transcription by interacting with proteins such as iron regulatory proteins. These proteins bind to specific structures within the 5′UTR, known as iron-responsive elements (IREs), thereby modulating translation in response to varying iron levels. [53, 54]. The 3′UTR is integral to the regulation of mRNA translation and its stability. In eukaryotes the elF4G interacts with poly (A)-binding proteins which bind to the poly (A) tail of the mRNA creating a circular loop, and this conformation is believed to promote efficiency [55]. It has been identified that 3′UTR plays a role in mRNA localisation as well [56].
It is known that some UTRs present in human genome promote higher translation rates than other UTRs. From a therapeutic standpoint, UTRs are key focus areas in research and development for maximizing translation rate and further stabilizing mRNA constructs. A study conducted by Asrani (2018) highlighted that combinations of different 5′UTRs of certain genes with 3′UTRs of other genes have introduced variation of expression in IVT mRNA. According to this study, the 5′UTRs of complement factor 3 (C3) and cytochrome p4502E1 (CYP2E1), consistently exhibit elevated expression of proteins in IVT mRNA [57]. Research conducted by Linares-Fernández et al. demonstrated that the 5′and 3′UTRs of ß-globin enhance translation efficiency. The study also indicated that using two 3′UTRs in tandem produces higher expression relative to using one ß-globin 3′UTR [58]. This effect of higher expression based on using repeated 3′UTR was later shown to be cell type-dependent and single 3′UTR has displayed higher protein expression that multiple tandem 3′UTRs [104]. The closely related field of bioinformatics has contributed to the strides made in UTR engineering via deep learning and in silico techniques. The development of novel UTRs contributes to higher expression and stability of mRNAs. Studies conducted by Cao et al. and Castilo-hair et al. show novel methods of developing optimized synthetic UTRs using in silico techniques, revolutionizing mRNA therapeutics [59, 60].
Poly(A) tail
The Poly(A) tail plays a critical role in translation initiation and the control of mRNA translation efficiency. According to the circular polysome model, translation initiation commences when eIF3 and met-tRNA bound 43s ribosome subunit forms a complex with 5′cap bound eIF4E and 3′poly(A) tail bound poly(A) binding protein (PABP) brought together by eIF4G. Translation efficiency has shown to be directly related to the length of the poly(A) tail. Most mammalian mRNAs consist of 200–250 adenine units in their poly(A) tail. Poly(A) tails also play a significant role in preserving the integrity of mRNA by reducing its length throughout the life span of the mRNA sequence. This reduction will continue until the length of the poly(A) tail has only 10–15 units left [54, 61–63]. Although this shortened poly(A) tail is unable to allow mRNA to associate with PABP or initiation factor eIF4E, preventing mRNA expression, the shortened tail associates with exoribonuclease 1 (XRN1), exoribonuclease RRP6, Dcp1p, Dcp2p and/or the cytoplasmic Lsm1p-7p followed by de-capping and subsequent degradation [64].
Several researchers have evaluated the effect of the length of poly(A) tail on translation efficiency. Peng et al., explored cell free in vitro translation system with Hela S3 cell cytoplasmic extract system in addition to in vitro translation using LM (tk-) cell transfection to investigate the translation efficiency relative to poly(A) tailing. This revealed that increase of poly(A) tail length from 0 to 98 adenosine residues progressively increased the translation from 1 to 25 fold [61]. According to Elango et al., increasing the poly(A) tail length of FLuc mRNA from 0 to 60 adenosine residues markedly increased FLuc expression in UMR-106 cells and this expression reduced after the length of 100 adenosine residues. Further findings in the study observed that mRNA translation was reduced for mRNA sequences containing heterologous sequences after the poly(A) tail [62].
The research conducted by Holtkemp et al. demonstrated that a length of 120 adenosines is optimal for translation in human dendritic cells [65]. This discovery was confirmed by Oh and Kessler, Avci-Adali et al. and Preskey et al. who have conducted studies of synthetic mRNA with poly (A) tail containing 120 adenosine residues where protein was successfully expressed in vitro [66–68]. S. Linares-Fernández et al. analysed variable of poly(A) tail lengths corresponding to their protein expression and observed that poly(A) tail containing 148 adenines increased the mRNA stability as well as expression in HeLa cells after 24 h post-transfection. However, the impact of poly(A) size has been recently re-evaluated, with recent studies demonstrating that longer tail (300 nucleotides) sequences are more optimal with novel mRNA designs [58]. The inconsistent research findings underscore the necessity for further investigation into the role of poly(A) tail length in mRNA kinetics. Grandi et al., examine the potential causes of these inconsistencies by decoupling mRNA degradation and translation in relation to poly(A) tail length. In this study, a library of identical mRNAs with varying poly(A) tail lengths was created, and mRNA kinetics were analysed in human embryonic kidney cells. The study demonstrates that while poly(A) tail length influences degradation, it does not directly determine the translation rate [69]. This research offers a potential explanation for the variation in translation efficiency associated with different poly(A) tail lengths. Recently Chen et al. developed a novel mRNA design with branched poly(A) tail. This multi-tailed mRNA encoded FLuc as the target protein while using RLuc (renilla luciferase) as the standard. HeLa cells were transfected using this mRNA and had displayed 4.7 to 19.5-fold increase in protein expression in 24, 48, and 72 h’ time intervals [70], showcasing the potential of promising developments focused on poly(A) tail structures in the mRNA design landscape.
Exogenous mRNA manufacturing
Before the COVID-19 pandemic, mRNA synthesis was a niche technology, and the scale of synthesis was relatively small. The COVID-19 pandemic saw a paradigm shift in the development and synthesis of mRNA for medical and research purposes. mRNA synthesis and manufacturing were upscaled to meet the surge in demand for the COVID vaccine. In 2021, mRNA synthesis became an urgent need due to the SARS-CoV-2 vaccine with a global demand of 1-2 billion doses. High volume exogenous synthesis of mRNA is mostly conducted using three methods: chemical, recombinant or enzyme-based systems [71].
Chemical synthesis (CS) of RNA utilizes phosphonamidite chemistry and solid phase support for continuous synthesizing RNA of less than 100 nucleotides (nt). The main limitation of solid phase chemical synthesis is size which is <100 nt. To synthesis longer RNA, short RNAs should be ligated together. This limits the scalability of this method of synthesis [71]. Yet CS has the distinct advantage of its ability to introduce site specific modifications like 2′-F modifications in to mRNA [72]. Abe et al. and Husseini et al. have developed a short mRNA therapeutic treatment known as minimal mRNA vaccine. According to Husseini et al., chemically synthesised minimal mRNA vaccine has shown promising results against melanoma with tumour inhibition and upregulation of cytokine expression after 22 days of treatment [73, 74].
Recombinant synthesis system (RSS) follows similar biomechanical method to recombinant protein production. In RSS, the mRNA sequence is amplified using either in vitro amplification or using in vivo amplification (with host cell like E.coli). Curry et al., has developed an in vivo mRNA synthesis platform using E.coli where E.coli are genetically engineered to use the bacteria’s mRNA synthesis system to synthesise synthetic mRNA. This novel platform has increased the product yield upto 40-folds compared to the unmodified E.coli expression systems [75]. RSS with in vivo amplification allows for the potential of large-scale production without a size limitation like CS, degradation of the transcribed mRNA by host nucleases and inability to use chemically modified nucleotides in the synthesis process are major draw backs of the system. Therefore, the enzyme-based system of in vitro transcription is considered as the gold standard method for the large-scale mRNA production [71, 76].
In vitro transcription (IVT) uses a linearized plasmid DNA (pDNA) template, or a DNA template amplified by polymerase chain reaction (PCR) for mRNA synthesis. The core enzyme that is used in this mechanism is bacteriophage RNA polymerase (T7, T3 or SP6), which recognizes the promoter in the DNA. Once the synthesis is completed, the dinucleotide m7GpppG as the structural homolog of endogenous Cap-50 and a poly(A) tail are added [17]. The main drawback of IVT system is the necessity of sequence-specific promoter for transcription initiation which limits terminal heterogeneity and the non-specific runoff in mRNA. Recent advancements in deep learning and sequencing has provided researchers with tools like synthetic libraries to optimize 5′ UTR sequences. Inclusion of human hydroxysteroid 17-beta dehydrogenase 4 genes’ 5′ UTR region in an mRNA vaccine candidate (CV2CoV), has enhances the translational capability of the mRNA [77]. Currently, IVT is the technology that is used for the synthesis of cost-effective large-scale clinically used mRNA following Good Manufacturing Practices (GMP) specification [66, 67, 71].
IVT system can be divided into three major stages: template amplification, in vitro transcription and purification. Depending on the scale (micro scale, bench scale, commercial scale) of mRNA synthesis, different techniques and equipment are employed based on the methods discussed previously [71]. In the micro scale synthesis, mRNA encoding linearized DNA is amplified using PCR, which is then used as the template for capped IVT reaction. The product is then purified using silicon-based columns such as Amicon Ultra-15 centrifugal filter units(30K membrane) (Millipore), RNeasy (Qiagen) and MEGAclear™ transcription clean-up kit (Thermofisher scientific) [66, 78]. At both bench and commercial scales, amplification involves producing mRNA from the DNA encoded in the plasmid, which is replicated using transformed bacterial cultures. On the bench scale, these cultures are typically grown using a batch processing protocol. In contrast, at the commercial scale, the transformed bacteria are propagated in bioreactors using a semi-batch processing approach. The extracted amplified template is used for IVT reaction, where the mRNA is synthesized and subsequently purified. A commonly used purification in bench scale is lithium chloride precipitation. On a commercial scale, techniques like ion exchange chromatography (IEC), ion-pair reverse chromatography (IPC), diafiltration using tangential flow filtration, and affinity chromatography are used. [3, 66, 71, 76]. The purification of modified mRNA is crucial for its functional activity. Contaminants present in in vitro transcribed RNA, such as double-stranded RNA, can induce immune responses, including the activation of type I interferons (IFNs) and proinflammatory cytokines. Therefore, the removal of these contaminants is imperative. The comprehensive study conducted by Karikó et al. demonstrates that high-performance liquid chromatography (HPLC) is one of the most effective purification methods. This technique yields mRNA that does not induce IFNs and inflammatory cytokines and is translated at levels 10- to 1000-fold higher in primary cells compared to poorly purified mRNA [78].
One drawback of using bacteria-based plasmid vectors for amplification is that researchers have noticed spontaneous deletions in the poly A tail. A study by Elong et al., tested different bacterial strains—DH5α, TOP10, GM2163, XL1-Blue, and XL10—to grow plasmids containing mRNA templates [62]. They found that all strains showed some reduction in poly A tail length, but XL1-Blue preserved the longest tail [62]. To improve the integrity of in vitro-transcribed (IVT) mRNA, scientists are developing new methods. He et al., created an advanced technique using a modified T7 polymerase and optimized IVT conditions. This approach achieved over 91% mRNA integrity, significantly improving on existing protocols [70].
mRNA encapsulation and delivery
Given the significance of mRNA and the first successful mRNA vaccine platform against COVID-19, the era of emerging disorders surged the escalating demand for targeted delivery systems for mRNA encapsulation and delivery [79–81]. As with other nucleic acids, mRNA is a macromolecule unable to cross evolutionary complex barriers (lipid bilayer and blood-brain barrier) making targeted delivery toward hard-to-reach cells a paramount challenge [82]. With the molecular weight of >106 g/mol, and electrostatic repulsion of negatively charged mRNA further impedes the penetration across the formidable cell membrane barrier [79, 82]. Moreover, without a delivery vehicle, the cell penetration potency of naked mRNA is lower [80]. Additionally, the shorter half-life together with enzymatic degradation by endonuclease, 5′and 3′exonucleases, raises further instability and immunogenicity concerns [80]. All these extracellular and intracellular barriers collectively necessitate establishing a delivery vector for the targeted release of mRNA in both in vitro (cell-based models) and (animal models) in vivo systems.
In past years, various physical methods have been deployed to facilitate mRNA uptake into the cells, primarily through transfection methods that introduce mRNA into the cytoplasm. Electroporation is one of the traditional methods with vast clinical applications including cancer immunotherapy, gene editing, and protein replacement. The principle of this method is based on nucleic acid delivery via enhanced membrane permeability through electric field induction [83]. Similarly, a gene-gun (a gold nanoparticle encapsulated mRNA vector) is one of the primitive mRNA delivery technologies for introducing mRNA into the targeted cell using a pressurized carrier gas [84]. Another method is microinjection-a type of mechanoporation that uses a micropipette/nanopipette for the direct injection of nucleic acid into intracellular space [85]. Microfluidic transfection is another innovative device for robust mRNA transport to chimeric antigen receptors expressing immune cells and NK cells. The mode of delivery is based on the principle of volume exchange for convective transfection (VECT) [86]. VECT devices are more advantageous over traditional electroporation platforms, especially for co-transgene expression, owing to their robustness, simplicity, and low genotoxicity [86].
Unfortunately, at present most cancer immunotherapies as well as prophylactic vaccines rely on local administration. With an emerging era of effective mRNA therapeutic delivery, the systemic route of administration is the major requirement but poses several targeted delivery challenges (Figure 6) [87, 88]. To circumvent this problem, encapsulation vectors-viral and non-viral-based systems have emerged harnessing the biomimetic potentials of nanobiotechnology [88]. Nonetheless, nanoparticles must be designed to mitigate the evolutionary cell barriers that hamper the targeted delivery of mRNA drugs to their main site of action. Upon systematic delivery, nanoparticle-based vehicles must escape endo-lysosomal vesicles, avoid renal clearance, and phagocytosis by circulating immune cells (Figure 6) [89]. They must also avoid non-specific plasma protein binding and be able to penetrate across-capillary endothelial barriers to reach their destination-the cytosol, for releasing the mRNA cargo for translation to occur [88]. Only about 2% of the nano-encapsulated drugs escape the endosomal entrapment, which has been incredibly challenging for macromolecules (>5 nm) to permeate the capillary endothelium; therefore, drugs are accumulated in the spleen and liver leading to unwanted toxicities. Finally, after reaching the dense extracellular matrix, the high osmotic pressure can impede fibrous proteins [90].
After reaching the extracellular matrix, the delivery vehicle must traverse a network of fibrous proteins and polysaccharides to reach the target cell membrane [1, 90]. Hence, the widespread clinical application of mRNA therapeutics faces a significant hurdle in finding safe and effective delivery systems.
Viral vs. non-viral vectors: While viral vectors like lentiviruses, adeno-associated viruses, and the Sendai virus have shown the capability to deliver nucleic acids systemically, their use can be hampered by unwanted immune responses [91, 92]. Importantly, many of these delivery materials have benefited from two decades of research of non-viral short-interfering RNA (siRNA) delivery [90]. Although siRNA differs from mRNA in being double-stranded, more rigid, and smaller (about 42 bases compared to over 1,000 bases for the average mRNA), they share the same chemical building blocks and require delivery to the same cellular destination, namely, the cytoplasm [90].
Non-viral vectors for mRNA delivery-clinical applications
The use of viral and synthetic vectors in nucleic acid delivery has revolutionized the field of genetic nanomedicine, notably contributing to the rapid development of highly effective COVID-19 vaccines (mRNA-based delivery platform) [7]. This breakthrough has opened new horizons for targeted nucleic acid delivery in nanomedicine. Both viral and synthetic vectors come with their own sets of advantages and disadvantages. Synthetic vectors offer the distinct advantage of limitless synthetic capabilities. Among nonviral vectors, lipid nanoparticles (LNPs), comprising four components, have emerged as the leading choice for mRNA delivery, notably utilized by Pfizer and Moderna in their COVID-19 vaccines [93].
Liposomes and lipid nanoparticles (LNPs)
Similarly, to circumvent the challenges of evolutionary barriers, several different non-viral mRNA delivery systems have emerged, with liposomes and lipid nanoparticles (LNPs) being prominent and long-standing examples [94]. Liposomes are spherical vesicles with one (unilamellar) or more (multilamellar) phospholipid bilayers encapsulating an aqueous core, with polar head groups, DOTMA (1,2-di-O-octadecenyl-3-trimethylammonium-propane), and DOTAP (1,2-dioleoyl-3-trimethylammonium propane), and zwitterionic DOPE (dioleoylphosphatidylethanolamine). The realm of these lipid-based formulations has been witnessed in several transfection agents such as Lipofectamine for a successful mRNA delivery, both in vitro and in vivo [94, 95]. Nevertheless, the major caveat of using cationic liposomes following the induction of inflammation is elicited by the interferon-γ response in animal models (mice) [96]. Moreover, cationic liposomes such as DOTMA and DOTAP are prone to anionic serum protein neutralization, affecting their therapeutic potential with toxicity concerns. Lipid nanoparticles differ from liposomes by composition and their formulation scheme is comprehensively discussed elsewhere. In general, are composed of polyethyleneglycol (PEG) modified lipids, cholesterol, ionizable cationionic lipids and phospholipids [96]. Like liposomes, LNPs following endocytosis can translocate mRNA into endolysosomal compartments, where mRNA is degraded. In the subsequent intracellular trafficking events, LNPs are pumped out of the cell via exocytosis, which further impedes effective cellular delivery [96]. To circumvent these issues, ionizable lipids (amphiphilic–positively charged at low pH and neutral at pH 7) have been developed as illustrated in Figure 5 [95]. These ionizable lipid systems imparted endosomal escape functionality to internalizing nucleic acid-based drugs in a proposed phenomenon known as the proton sponge effect [97]. Although the exact mechanism is not known, it has been proposed that the positively charged LNP component facilitates interaction with the negatively charged endosomal membrane leading to ATP-driven acidification. These events follow a gradual shift in the pH from five to 6, rendering the endosomal membrane leaky by disrupting the membrane lipid bilayer. The details of this hypothesized phenomenon are reviewed comprehensively in previous reviews [97]. Resultingly, the nucleic acid cargo escapes the endosomal entrapment and is targeted toward its site of action. On the targeting front, further studies need to focus on developing RNA-based therapeutics using new targeting material and chemistries with enhanced endosomal escape properties [82, 95, 97].
FIGURE 5
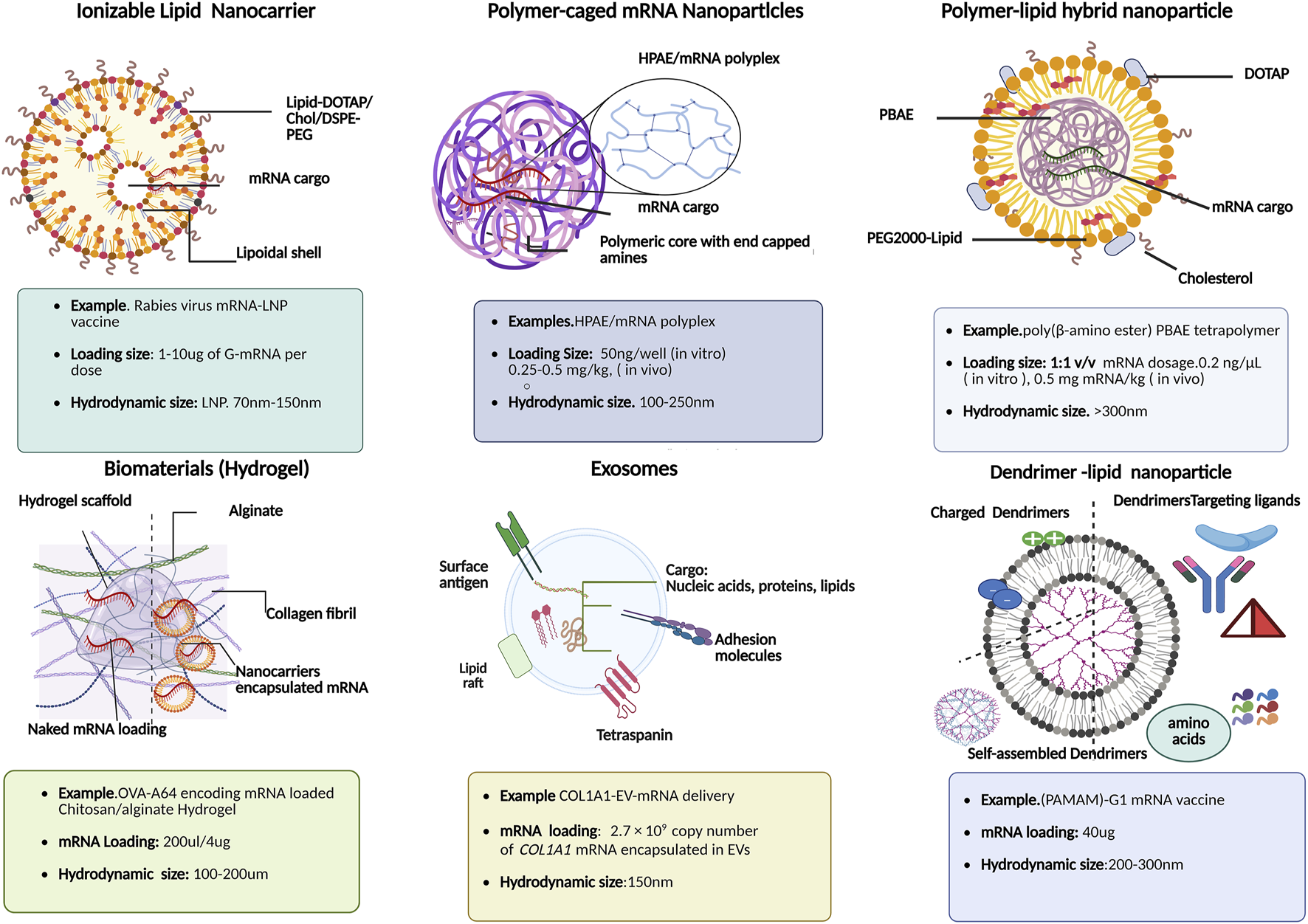
Schematic illustration of various functional mRNA-loaded nanocarriers: their composition and physiochemical features for downstream clinical applications in mRNA therapies.
Interestingly, one of the unique features of lipid nano formulations amenable to inefficient mRNA loading is a balanced ratio of ionizable lipids, and helper lipids such as 1,2-Dioleoyl-sn-glycero-3-PE (DOPE) and distearoylphosphatidylcholine (DSPC), which promote endosomal membrane fusion, polyethylene glycol (PEG) for improved bioavailability by reducing plasma protein binding, promoting cell-specific interactions toward non-hepatic organs, especially lymph nodes [98]. Besides translation outcomes of LNP-based mRNA delivery systems in protein replacement therapies, emerging mRNA-based vaccines are in the limelight for their role in cancer immunotherapies as rabies virus glycoprotein (RABV-G) mRNA-LNP based vaccines as shown in Figure 5. It is worth noting that the mRNA loading size in rabies virus LNP based vaccination varies from 1 to 15 µg per dose. For instance, previous studies elucidated sharp rise in humoral immune response in in vivo studies using range of dosages (0.1, 0.3, 3, and 10 µg) of RABV-G-mRNA; the high dose in vivo models of BALB/c mice with, 3 μg dose afforded maximum RABV-G-specific antibody titres (20.3 IU/mL, week 4) with single intramuscular (IM) injection [99–101]. In the subsequent immune response kinetic studies, it was revealed that a further 10 μg booster vaccination could increase and sustain long-term immune response kinetics with stable rise in Rabies virus neutralizing titres (1000–2000 IU/mL) in LNP formulation vs. empty LNPs (negative control) for up to 35 weeks [102]. Furthermore, lipid nanoformulation with helper lipids [e.g., cholesterol, dioleoylphosphatidylethanolamine (DOPE), dioleoylphosphocholine (DOPC), distearoylphosphatidylcholine (DSPC), and 1,2-Dimyristoyl-rac-glycerol (DMG-PEG) can further enhance the delivery efficacy by providing shielding effects and thus, stability against RNases and endonucleases within the cytosol and plasma membrane [103].
Thus, both lipoplexes and lipid nanoparticle-based mRNA formulations (Figure 5) and (Table 2) have already entered various stages of clinical trials for delivery toward extrahepatic tissues, seem promising, and have already broadened the horizon of RNA nanotechnology platforms.
Cationic polymeric nanoparticles
In the emerging era of nanoengineering, polymeric-based delivery vectors such as polymer-lipid hybrid nanoparticles, polymeric caged nanoparticles, dendrimer based lipid nanoparticles, etc., are some of the stand-out examples of non-viral RNA delivery systems, due to their inherent biocompatible and biodegradable nature [103]. In recent years poly(β-amino esters) (PBAE) has emerged in mRNA-targeted delivery to various parenchymal cells including pulmonary immune cells. For instance, OMPBAEs, an oligopeptide-modified PBAE, are designed for liver-specific targeting using mRNA transfection. Similarly, hyperbranched derivatives of PBAE maximized the mRNA delivery toward lung epithelium, shown in Figure 6 as polymer-caged mRNA nanovectors [104]. Moreover, in addition to terminal amino acids chemistries during formulation scheme, the mRNA loading efficiency and release kinetics is further influenced by polyplex size. For example, smaller sized (100–250 nm), biodegradable highly branched PBAE/mRNA complexes (Figure 5) showed high (80%) mRNA binding efficiency compared to larger (∼2 μm) counterparts [104]. In addition, to treat liver disorders, studies reported dendrimer-modified LNP systems used as promising theragnostics. For example, PEGylated BODIPY dyes to design PDB lipids for mRNA vaccine delivery to hepatic cells revealed therapeutic potential in liver cancers. Similarly, researchers used self-replicating and adjuvant-free mRNA vaccines against life-threatening infectious diseases (H1N1 influenza, Zika, Ebola, and Toxoplasma gondii) which made use of biodegradable polypropylene imine-based dendrimers or polyamidoamine (PAMAM-G1 mRNA vaccine) as shown in Figure 5 [105, 106].
FIGURE 6
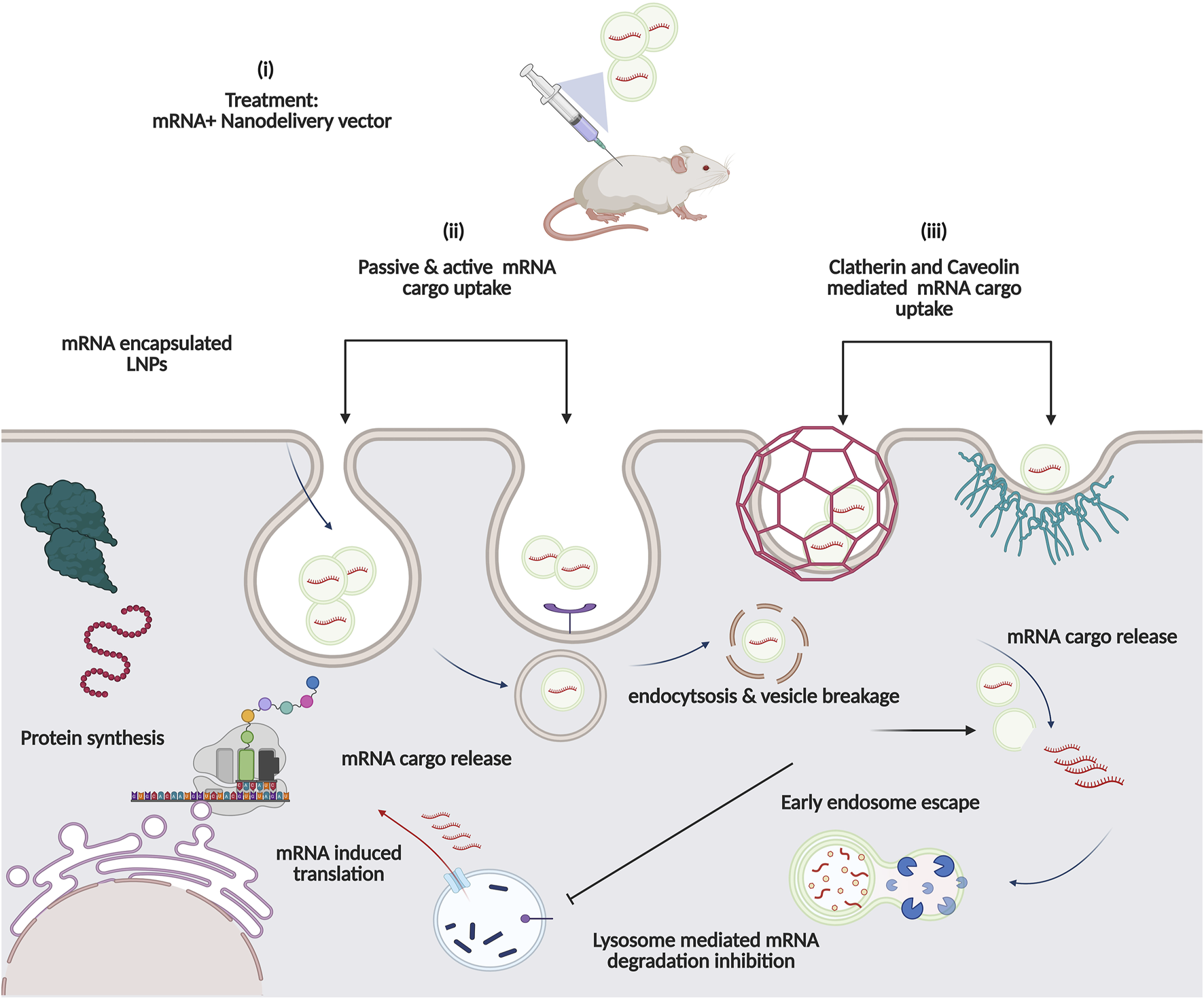
Proposed Intracellular pathway of Nanocarrier-based mRNA drugs: (i) In vivo immunisation with mRNA-based Nanovector formulation. Cell uptake of mRNA cargo occurs via either (ii) active (receptor-mediated) or passive diffusion across lipid bilayer or (iii) Clatherin and caveolin Mediated. Enhanced delivery with endolysosomal escape functionality aids in robust protein expression.
Another category of intracellular nucleic acid delivery system is based on cell-penetrating peptides, which deliver mRNA cargo via various proposed mechanisms, i.e., either through lateral or passive diffusion penetrating the membrane barrier, involving clatherin or caveolin mediated cell uptake (Figure 6) [105, 106].
Exosomes
Nevertheless, despite the emergence of these ascribed mRNA delivery systems, the poor stability and immunogenicity are still major drawbacks, seriously hampering their clinical utility. Moreover, due to the instability of naked mRNA, various protein replacement therapies have been regarded as an alternative strategy but cause highly immunogenic responses. The intrinsic properties of exosomes such as the “homing effect” make these biological nanocarriers an efficient and safe mRNA delivery vector [107]. First discovered in the 1980s, exosomes can transport proteins, nucleic acids (miRNAs and mRNAs), vitamins, and other intracellular cargo toward their target destination through signal transduction and cell-cell communication (Figure 5) [107, 108]. Previously, exosomes have been exploited to deliver mRNA toward immune cells to establish robust immunity against COVID-19 [107, 109]. These biologically derived extracellular vehicles (EVs) can target tumour cells by penetrating across the blood-brain barrier, making them an iconic mRNA-packing vector for brain targeting and other personalized therapies [110]. Yang explored in vivo, exosome-mRNA delivery which also enhanced survival rate with a significant reduction in tumour growth by targeting a tumour suppressor gene- Phosphatase and tensin homolog in PTEN-knockdown glioma mice [110].
Similarly, mRNA-encapsulated exosomes have also been explored as a promising collagen replacement therapy. For instance, exosomes derived from human dermal fibroblast were engineered with full length (4000 nt) long extracellular matrix alpha 1 collagen (COL1A1) mRNA (Figure 5). It was fascinating to discover that intradermal COL1A1-EV-mRNA delivery reduced dermal wrinkles in a photoaged UV irradiation mice model [111]. Additionally, inhalable EV-mRNA has shown robust delivery with higher efficiency compared to liposomes-mRNA delivery vectors [112].
Despite the outstanding characteristics of exosome-based mRNA delivery systems over LNPs, the challenges of using exosomes such as large-scale isolation and purification for subsequent clinical applications remain a paramount setback [107, 113]. The existing methods are not only costly but also generate low exosome yield. Furthermore, often using conventional methods like size-exclusion, immunoaffinity, ultrafiltration, etc. encounter contamination problems (with proteins or non-exosome vesicles) making the separation of therapeutically functional exosomes more tangible.
Immunogenicity and toxicity of exosomes have been the additional bottleneck in the design and optimization of efficient exosome-mRNA delivery systems [114]. Therefore, to have a must-have ‘mighty RNA delivery system,’ it is essential to adhere to GMP while selecting the most optimal and efficient exosome engineering method [115]. Understanding and determining vital cell sources for therapeutically functional exosomes is one of the first crucial steps to mitigate any contaminations and would potentially facilitate selective and efficient loading of mRNA cargo on these biologically active extracellular vesicles (Figure 5) [116]. In the future, it is anticipated that the choice of appropriate quality control measures together with standard protocols for production and characterization will be taken into consideration to generate reproducible and clinically functional exosomes [117].
Hydrogels
In recent years, researchers have explored multifaceted role and drug release kinetics of variety of biomaterials loaded RNA [118, 119]. Hence, owing to good biocompatibility, and better biodegradability than LNP counterparts, hydrogels are one such biomaterial which have emerged as a promising injectable/implantable delivery platform for sustainable (burst/pulsating) RNA release. The tunable physiochemical features of hydrogels further enable the retention of functional mRNA activity in biomaterials-based immunotherapies [118, 120–122]. Hydrogels are three-dimensional porous and permeable scaffolds composed of either natural (chitosan/alginate), synthetic polymers, i.e., PEI, or even hybrid polymer complexes (hyaluronic acid and polyvinyl alcohol; HA.PVC) [119]. Furthermore, the robust design scheme of RNA-encased hydrogel platforms (Figure 5) depends on the nature of the loading cargo-whether a naked RNA cargo (siRNA, miRNA, mRNA) or additional nanocarriers (organic and inorganic nanoparticles, lipid or polymeric-based nano vectors) have been packaged onto hydrogel matrix. Thus, for effective localized delivery of naked RNAs with minimal off-target toxicities, the type of RNA-hydrogen interactions viz, covalent, electrostatic, hydrophobic, non-specific interactions or combination, play a significant role in regulating “on-demand” release profile, retention time, RNA stability with minimal off-target toxicities [118, 119]. A comparison of these conjugation strategies with technical limitations for efficient RNA packing is detailed comprehensively elsewhere [119]. Similarly, Yan et al. explored injectable chitosan/alginate hydrogel scaffolds (Figure 5) for rapid and controlled delivery of mRNA lipoplex (mRNA-OVA-A64 vaccine) in vivo (C578L/6J mice) [120]. Furthermore, in subsequent experiments, luciferase gene expression corresponding to IgG humoral response was found to be five-fold higher than the systematic injection [6]. Interestingly, stimuli response hydrogel-based RNA carriers (active or passive) have also been reported and engineered to modulate further the “on-demand/pulsating” RNA release profile, a crucial variable that affects downstream targeted RNA delivery approach. For instance, enzyme-responsive (matrix metallopeptidase MMP-2 degradable hydrogel), pH-responsive (DNA nanohydrogels for mRNA delivery), and Light sensitive hydrogels (UV-cleavable Chol-miR-26a) are among the proposed stimuli-responsive hydrogels utilized in “active” RNA cargo delivery, which were previously discussed [118, 119].
Rwandamuriye et al. have most recently engineered a hyaluronic acid-based hydrogel encapsulating an immunostimulant-a polyinosinic:polycytidylic acid poly(I:C) (a toll-like receptor 3-TLR3 agonist). Collectively, physiochemical characterizations including scanning electron microscopy, quantitative micro elastography, and Young’s Modulus confirmed the size, morphology as well as the surgically optimal hydrogel formulation (2.5% w/v HA and 3.5 mol% DTPH crosslinker) respectively, for safe and effective downstream biomedical applications. The resulting poly(I:C)-hydrogel optimized in surgical tumour model revealed promising outcomes in intraoperative immunotherapy, i.e., improved recurrence free-survival post-surgery, with transient alpha (IFNα) response rate and improved tumour sensitivity toward ICT tested mouse model (C57Bl/6J models). The safety and immunogenicity (type I IFN dependent responses) as well as the delivery efficacy of loaded immune adjuvants [poly(I:C)) in a proposed biomaterial matrix were further validated in canine veterinary models with cancer [121, 122]. The complete protocol guide on systematic “debulking surgery and bandaging method” of C57Bl/6J models followed by subcutaneous injection of poly(I:C) based hydrogels at tumour resection site are comprehensively explained in detail and published elsewhere [122].
Collectively, researchers proposed a robust alternative to neo-adjuvant and adjuvant immunotherapies which are only limited to certain cancer therapies, thus offering a facile advancement of exploiting low and prolonged RNA immunotherapeutic loaded hydrogels delivery vectors with broad spectrum biomedical applications from wound healing to anticancer and cardioprotective function against myocardial infraction.
mRNA-based therapeutic strategies and applications
In general, eukaryotic mRNA translation initiation can be divided into three stages. Firstly, 40S ribosome subunit binds to mRNA in the vicinity of 5′Cap with the assistance of several eukaryotic initiation factors (elFs). Subsequently, the 40S ribosome scans through the 5′UTR to find the initiation codon, and eventually, 60S ribosomal subunit joins the 40S subunit releasing the elFs. As a result, 80S ribosome is formed and starts the process of elongation [52]. Unlike in vivo synthesised mRNA, synthetic mRNA enters the cell from extracellular space; yet both display their activity in cytoplasm without integrating to the genomic DNA (Table 1). Using host machinery, IVT mRNA is translated into peptide which undergoes post-transcriptional modification to become bioactive protein. IVT mRNA is designed to resemble the natural post-modified mRNA in the cytoplasm [123]. mRNA-based therapeutics can be divided into three main therapeutic strategies: cellular modulation, protein replacement, and immunomodulation. The cellular modulation and protein replacement therapeutic strategies involve mRNAs that encode a functional protein, which can be expressed in the target cell as an endogenous protein, a functional foreign protein or proteins used for gene editing or cellular modulation (Figure 7; Tables 1, 2). In the context of mRNA-based immunomodulation, the introduced mRNA expresses antigens that trigger immune responses, either cell-mediated or antibody-mediated, against an external pathogen or a tumour cell [104].
TABLE 1
| Therapeutic strategies | Applications | References |
|---|---|---|
| mRNA based protein replacement | Cardiac, collagen replacement, treatments for rare genetic disorder | [12, 17, 104, 111] |
| mRNA based immunomodulation | mRNA encoded monoclonal antibody therapy | [104, 125, 126] |
| mRNA- based CD4+ T cell therapeutics | [104, 127] | |
| mRNA-based T lymphocyte therapeutics (editing primary T cells) | [17, 104, 128] | |
| mRNA based cellular modulation | Induced pluripotent stem (iPS) therapeutics Gene editing | [17, 68, 104, 129] |
mRNA therapeutic Strategies.
TABLE 2
| Therapeutic strategy | Disease or condition | NCT no. | Candidate name | Molecular target | Delivery vehicle | Administering method | Sponsor | Development stage | References |
|---|---|---|---|---|---|---|---|---|---|
| mRNA based immunomodulation | COVID-19 | NCT04860297 | mRNA-1273 | S-2P | LNP | Intramuscular injection | ModernaTX, Inc | III | [205] |
| NCT04816669 | BNT162b2 | S-2P | LNP | Intramuscular injection | BioNTech SE | III EUA & CMA | [206] | ||
| NCT04523571 | BNT162b1 | Trimerized RBD | LNP | Intramuscular injection | BioNTech SE | I | [207, 208] | ||
| NCT04847102 | ARCoV | RBD | LNP | Intramuscular injection | Walvax Biotechnology Co., Ltd. | III | [209] | ||
| NCT04668339 | ARCT-021 | Naive S protein | LNP/ | Intramuscular injection | Arcturus Therapeutics, Inc. | II | [210] | ||
| NCT04798027 | MRT5500 | S-2P with a furin cleavagesite mutant | LNP | Intramuscular injection | Sanofi Pasteur, a Sanofi Company | I/II | [211] | ||
| NCT04566276 | ChulaCov19 | Transmembrane S protein | LNP | Intramuscular injection | Chulalongkorn University | I/II | [212, 213] | ||
| Rabies | NCT02241135 | CV7201 | Rabies virus glycoprotein (RBVG protein) | Protamine-condensed mRNA | Intradermal injection | CureVac | I | [214, 215] | |
| NCT03713086 | CV7202 | RBVG protein | LNP | Intramuscular | CureVac | I | [216, 217] | ||
| Influenza | NCT04956575 | mRNA-1010 | Hemagglutinin (HA) from 4 influenza virus strains (influenza A (H1N1) Influenza A (H3N2) Influenza B (Yamagata lineage) Influenza B (Victoria lineage)) | LNP | Intramuscular injection | ModernaTX, Inc. | I/II | [218] | |
| NCT05333289 | mRNA-1020 and mRNA-1030 | Haemagglutinin and neuraminidase to conserved regions of the virus | LNP | Intramuscular injection | ModernaTX, Inc. | I/II | [219, 220] | ||
| NCT06431607 | GSK4382276A | HA from influenza virus antigens (H1N1, H3N2, Yamagata lineage, Victoria lineage) | LNP | Intramuscular injection | GlaxoSmithKline | II | [221] | ||
| NCT05823974 | LNP | Intramuscular injection | GlaxoSmithKline | I/II | [222] | ||||
| Chikungunya virus infection | NCT03829384 | mRNA-1944 | Anti-Chikungunya antibody | LNP | Intravenous infusion | ModernaTX, Inc. | I | [223] | |
| HIV infection | NCT05217641 | HVTN 302 | Stabilized HIV envelope (Env) trimers Native-like HIV-1 Env trimers (BG505 MD39.3, BG505 MD39.3 gp151 and BG505 MD39.3 gp151 CD4KO) | LNP | Intramuscular injection | National Institute of Allergy and Infectious Diseases (NIAID) | i | [224] | |
| NCT05001373 | IAVI G002 | eOD-GT8 60mer Env (mRNA-1644) Core-g28v2 60mer Env (mRNA-1644v2) | LNP | Intramuscular injection | International AIDS vaccine initiative | I | [225] | ||
| NCT05903339 | HIV-1 ferritin nanoparticle vaccine | Native-like HIV-1 envelope trimer | LNP | Intramuscular injection | National Institute of Allergy and Infectious Diseases (NIAID) | I | [226] | ||
| Cytomegalovirus infection | NCT03382405 | mRNA-1647/1443 | CMV glycoprotein H (gH) pentamer complex | LNP | Intradermal | ModernaTX, Inc. | I | [227–229] | |
| Herpes Zoster (HZ) | NCT05701800 | mRNA-1468 | Glycoprotein E | _ | Intramuscular | ModernaTX, Inc. | I/II | [230] | |
| NCT05703607 | BNT167 | Glycoprotein E | _ | Intramuscular | Pfizer | II | [231] | ||
| Human metapneumovirus and parainfluenza infection | NCT03392389 | mRNA-1653 | Human metapneumovirus and human parainfluenza virus type 3 vaccine | Lipid nanocarrier | Intradermal | Moderna | I | [232, 233] | |
| Melanoma | NCT01676779 | TriMixDC-MEL | MAGE- A3, MAGE-C2, tyrosinase and gp100 | DCs | Intradermal injection | Universitair Ziekenhuis Brussel | I | [234, 235] | |
| NCT04526899 | BNT111 | NY-ESO-1,tyrosine,MAGE-A3, and TPTE) | Lipoplex | Intravenous | BioNTech | II | [236, 237] | ||
| Triple negative breast cancer | NCT02316457 | [1] 3 TAAs selected from a warehouse and p53 RNA; [2] Neo-Ag based on NGS screening | Lipo-MERIT, DOTMA(DOTAP)/DOPE lipoplex | Intravascular | BioNTech | I | [238] | ||
| NSCLC | NCT03164772 | CV9202 | NY-ESO-1, MAGE-C1, MAGE-C2, survivin, 5T4, and MUC1 | Protamine-condensed mRNA | Intradermal injection | Ludwig institute for cancer research | I/II | [239, 240] | |
| NCT03948763 | - | K-ras encoding mutation (mRNA-5671) | Lipid nanocarrier | Intramuscular | Merck sharp &Dohme | I | [241, 242] | ||
| Colorectal cancer | NCT04486378 | RO7198457 | Personalised neoantigen | Lipid nanocarrier | Intravascular | BioNTech | II | [243] | |
| Head and neck cancer | NCT04534205 | BNT113 | E6 & E7 oncoproteins of HPV16 | Lipoplex | Intravenous | BioNTech SE | II | [244–246] | |
| Solid Tumours | NCT04503278 | BNT211-01 | Claudin 6 (CLDN6) chimeric antigen receptor T cells (CAR-T) | Lipoplex | Intravenous | BioNtech & gene Therapies | I | [247, 248] | |
| NCT03313778 | mRNA-4157 | Neo- Ag | Lipid nanocarrier | Intradermal | Moderna | I | [249–251] | ||
| High-risk melanoma | NCT05933577 | mRNA-4157 (V940) with pembrolizumab MK-3475 | Merck sharp & Dohme LLC. | III | [252] | ||||
| Non-small cell lung cancer | NCT06077760 | Merck sharp & Dohme LLC. | III | [253] | |||||
| Pancreatic cancer | NCT04161755 | _ | Personalise neo-antigen (neo-Ag) | Lipoplex | Intravenous | Memorial Sloan Kettering Cancer Centre, USA | I | [254, 255] | |
| Autologous cancer | NCT03480152 | mRNA 4650 | Neo-Ag mRNA | LNP | Intramuscular | National Cancer Institute (NCI) | I/II | [256, 257] | |
| Methylmalonic acidaemia | NCT03810690 | mRNA-3704 | Methylmalonyl-CoA mutase | Unknown LNP | Intravenous injection | ModernaTX, Inc. | I/II (Withdrawn) | [258] | |
| mRNA based protein replacement therapy | Propionic acidaemia | NCT04159103 | mRNA-3927 | Propionyl-CoA carboxylase | Unknown LNP | Intravenous injection | ModernaTX, Inc. | I/II | [259] |
| Ornithine transcarbamylase deficiency (OTD) | NCT05526066 | ARCT-810 | Ornithine transcarbamylase | LNP | Intravenous injection | Arcturus Therapeutics, Inc. | II (ongoing) | [260, 261] | |
| Cystic fibrosis | NCT05668741 | VX-522 | CFTR | LNP | Inhalation | Vertex pharmaceuticals incorporated | I/II | [262, 263] | |
| NCT03375047 | MRT5005 | CFTCR | LNP | Inhalation | Translate Bio, Inc | I/II | [264, 265] | ||
| Type II diabetes | NCT02935712 | AZD8601 | VEGFA | Nake mRNA | Intradermal injection | AstraZeneca | I (completed) | [266, 267] | |
| Heart failure | NCT03370887 | AZD8601 EPICCURE | VEGFA | Naked mRNA | Epicardial injection | AstraZeneca | II (completed) | [268–270] | |
| mRNA based cellular modulation | Transthyretin amyloidosis with polyneuropathy (TAP) | NCT04601051 | NTLA-2001 | CRISPR–Cas9 | LNP | Intravenous injection | Intellia Therapeutics | I | [271, 272] |
| NCT06128629 | III | [273] | |||||||
| Hereditary transthyretin amyloidosis | NCT04601051 | NTLA-2001 | CRISPR-Cas9–based transthyretin (TTR) editing | Lipid nanocarrier | Intravascular | Intellia Therapeutics | I | [272, 274] |
mRNA therapeutic development.
FIGURE 7
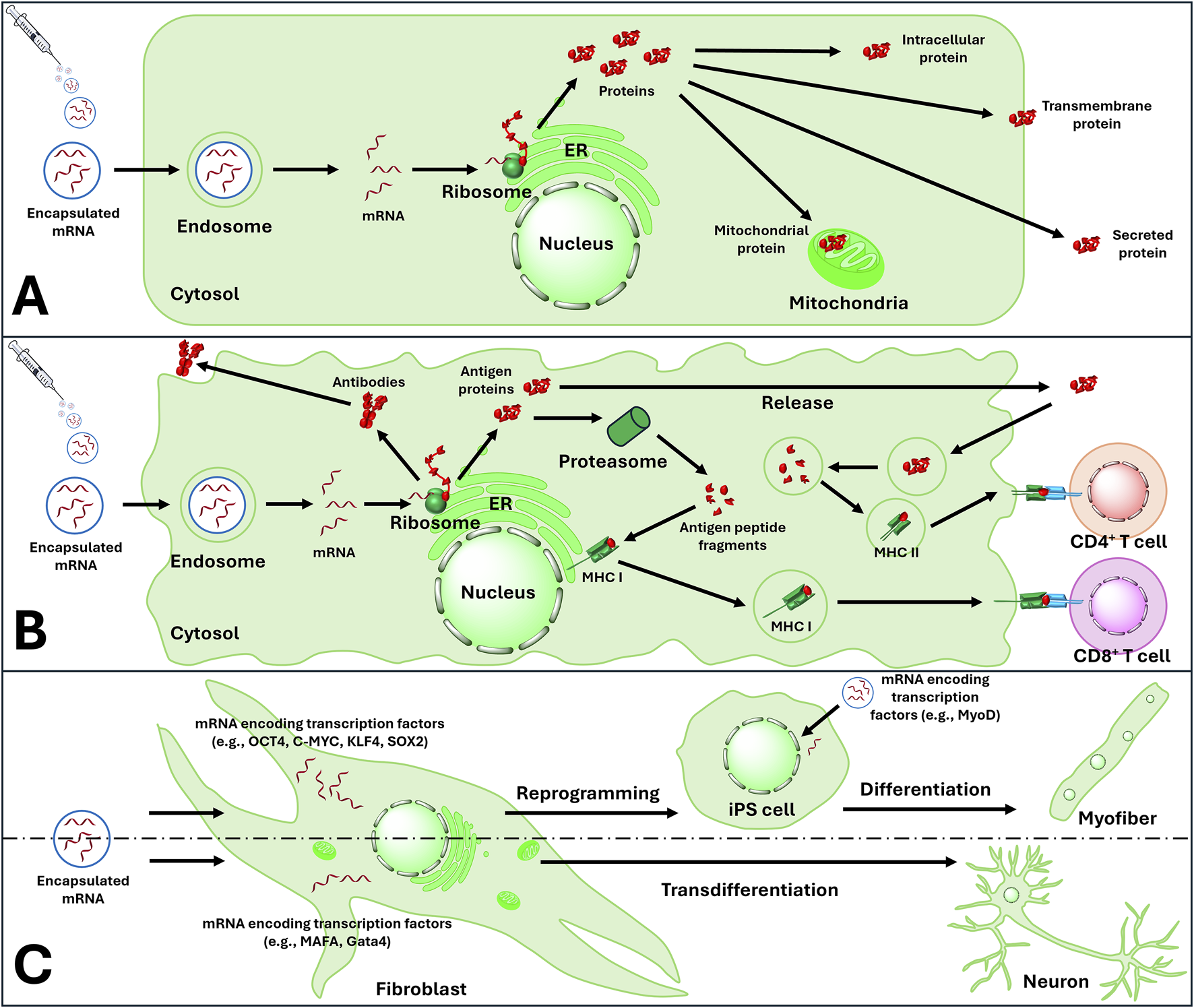
mRNA based therapeutic strategies include: (A) protein replacement therapy (B) Modulation of immunity by activating active and passive immunity and (C) reprogramming and transdifferentiation of audlt cells. ER, endoplasmic reticulum; MHC I, major histocompatibility complex class I; MHC II, major histocompatibility complex class II; CD4, cluster of differentiation 4; CD8, cluster of differentiation 8; iPS cells, induced pluripotent stem cells.
Since mRNA’s discovery, researchers have envisioned its therapeutic potential. SARS-CoV-2 prompted extensive resources for mRNA-based treatments and vaccines, yielding technological breakthroughs. These advances reveal synthetic mRNA’s therapeutic possibilities, warranting thorough patent examination [124].
mRNA-based protein replacement
mRNA based protein replacement therapy (PRT) is defined as treatment that increases the concentration of a specific protein that has been deficient in a cell due to absent or incomplete synthesis because of a genetic mutation. mRNA possesses the capacity to encode the protein that is deficient in the patient [12]. Therefore, mRNA-based protein replacement has less risks in mutagenesis. mRNA designed and synthesised with protein coding region of the target gene incorporating modified nucleotides, has shown to synthesize proteins in mouse models with low immunogenicity, prolonged stability and high efficiency [17]. Following these principal examples, we explore the innovative patented therapies applying IVT mRNA as protein replacement and cellular modulation therapy.
Citrullinemia type 2 (OMIM ID 603471; 603859)
Citrullinemia type 2 (CTLN2) or adult-onset type 2 citrullinemia, is a class of urea cycle disorders, inherited as an autosomal recessive trait [130]. CTLN2 is characterized by markedly high levels of L-citrulline in both the liver and blood of affected patients with a severe Citrin deficiency (CD) [130–134]. Epidemiological studies have shown that prevalence rates for pathogenic bi-allelic variants are commonly found in East Asian populations; mainly Japan with an incidence rate of 1:69 and 1:65 in China [135, 136]. The occurrence rate of CTLN2-induced Citrin deficiency was most recently reported in France, UK and Canada, which implies that CD has now emerged as a pan-ethnic disease worldwide [135, 136].
The genetic studies elucidated that the underlying cause of Citrin deficiency is due to the mutation in SLC25A13-a mitochondrial localized liver-specific aspartateS/glutamate transporter encoding gene for citrin protein, located on 7q21.3 locus [133, 136, 137]. The citrin/aspartate/glutamate carrier isoform 2 (AGC2) plays a critical role in hepatocyte glycolysis by providing a mitochondria-specific energy shuttle in the form of Ca+ dependent malate-aspartate nicotinamide adenine dinucleotide hydrogen (NADH) carrier [131, 135]. In citrin-deficient individuals, without NADH, ammonia is unable to be removed during the urea cycle leading to symptoms of hyperammonia [135]. The metabolic pathways involving de novo lipogenesis, fatty liver-specifically non-alcoholic fatty liver disease (NAFLD) and hyperlipidemia have been well documented in scientific literature [131, 132, 135, 136, 138].
Given the severity of the disease manifestation in CD patients with severe hyperammonemia conditions, liver transplantation is generally the last resort to manage the clinical pathology of CTLN2 [132, 139–142]. Presently, recommended dietary therapies for adult-onset type II citrin deficiency are mostly main chain triacylglycerol (MCT) oils (5 mL twice daily), Sodium pyruvate (0.1–0.3 g/kg/day), low carbohydrates and high protein supplements [132, 133, 140, 142]. In practice, patient compliance, recorded poor efficacy and reported side effects (carbohydrate toxicity) have lessened the effectiveness of traditional treatment methods [131, 132, 134, 139, 142]. Treatment regimens mentioned earlier has succeeded in the complete management of inherited metabolic CTLN2 disorder [131, 136, 139].
In 2019, Cao developed a safe and effective mRNA therapy based on LNPs, bearing a codon optimised and chemically modified mRNA variants for human citrin (citrin-mRNAcov) [134]. Subsequent intravenous (IV) administration to both mammalian liver cell-lines (HepG2) in vitro and CTLN2 mouse model in vivo corrected the subcellular localisation and expression of a fully functional citrin protein in liver mitochondria, i.e., almost equivalent to 2–5% of wild-type expression levels [134]. Accordingly, in 2022, Martini (US patent NO. 20220071915A1) designed intracellular mRNA-lipid nanoparticle-based delivery technology encoding a full functional fragment of citrin protein, sharing 85% sequence homology to wild-type human Citrin Isoform 2 (Seq ID No. 3) [136]. Furthermore, the designed mRNA-based LNP technology further addressed the hepatic citrin levels (increased by 50%), a key biomarker for fatty liver in citrullinemia type 2 patients. The hepatic citrin remained at optimal levels for 24–96 h post-administration [136].
Acute intermittent porphyria (OMIM ID 176000)
Acute intermittent porphyria (AIP, also known as Swedish porphyria) is a rare metabolic disease that is characterized by deficient heme biosynthesis and inherited in autosomal dominant manner [143, 144]. AIP occurs because of mutations in the gene encoding porphobilinogen deaminase (PBGD), also known as hydroxymethylbilane synthase (HMBS). The morbidity of AIP is 1 in 20,000 [145], and more than 200 pathogenic mutations have been recognized [143]. Defects of PBGD lead to insufficient heme biosynthesis, which stimulates upregulation of δ-aminolevulinic acid synthase (ALAS) and eventually accumulates neurotoxic metabolic intermediates ALA and PBG [146–150]. Intravenous heme administration is a useful treatment for acute attacks, as it suppresses the physiological feedback of insufficient heme storage, thus reducing the production of ALA and PBG, because of ALAS inhibition. Liver transplantation is the definitive solution for AIP patients [151–153]. mRNA encoding PBGD for the treatment of AIP has been patented [154], and this strategy not only restores the expression of PBGD and its activity but also decreases the level of the neurotoxic metabolites (ALA and PBG). Notably, intravenous administration of 0.5 mg/kg PBGD mRNA formulated by lipid nanoparticles into Cynomolgus macaque achieved ∼40% and ∼90% increases in the activity and expression level of PBGD, respectively, in comparison to the phosphate buffered solution (PBS) treated animals [154].
Argininosuccinate synthetase deficiency (OMIM ID 215700)
Arginosuccinate synthetase deficiency (ASD) also known as argininosuccinic acid synthase deficiency and citrullinemia type I, is a urea cycle disorder (UCD). In UCDs, ammonia (nitrogenous waste of protein metabolism) fails to be converted into urea and released through urine. The resultant accumulation of ammonia in the body causes toxic effects. Arginosuccinate synthetase is a key enzyme in urea cycle, and deficiency of this enzyme leads to inability to catalyze the formation of argininosuccinic acid. This process results in the buildup of upstream substrates, specifically citrulline and aspartic acid, which eventually leads to UCD and hyperammonemia. ASD is inherited in an autosomal recessive pattern and is attributed to mutations in the ASS1 gene responsible for encoding arginosuccinate synthetase [155, 156]. The prevalence of ASD is ∼1/57,000 [157], and for patients with ASD, nutritional management is essential in preventing complications of hyperammonemia [158]. An mRNA therapy for ASD has been patented [159], providing an mRNA encoding arginosuccinate synthetase that is able to restore sustained and highly efficient in vivo synthesis of the enzyme, when delivered in lipid nanoparticles. Furthermore, administration of the mRNA (1.0 mg/kg) to ASS1 knockout mice achieved ∼50% reduction of ammonia in plasma on the following day [159].
Cystic fibrosis (OMIM ID 219700)
Cystic fibrosis (CF) is an autosomal recessive genetic disorder caused by pathogenic variants of the gene encoding cystic fibrosis transmembrane conductance regulator (CFTR) [160–162]. CFTR is a transmembrane protein that forms a chloride channel. Mutations in the CFTR gene lead to synthesis of defective proteins that are unable to carry out chloride ion transport or even cannot reach the cell membrane. The resultant accumulation of chloride ions leads to generation of thick secretions (mucus) in different organs such as lungs and liver [163]. Patients with CF eventually suffer from loss of lung function as a result of progressive lung disease [164, 165]. Currently there are ∼89,000 people affected by CF worldwide [165]. First-line treatments for the CF-associated lung disease consist of administration of mucolytics (mucus thinner), anti-inflammatories, airway clearance, and antibiotics. Small molecular CFTR modulators such as ivacaftor have been approved by regulatory bodies as well [165]. An mRNA treatment for CF has been patented [166], providing an CFTR mRNA encapsulated in lipid nanoparticles that can increase the expression level and/or activity of CFTR in subjects, thus reducing the production of toxic metabolites associated with CFTR deficiency and/or dysfunction [166].
Fabry disease (OMIM ID 301500)
Fabry disease is a progressive, X-linked inherited lysosomal storage disorder (LSD), characterized by deficient activity of lysosomal α-galactosidase A (α-Gal A), affecting glycosphingolipid metabolism [167–170]. α-Gal A is coded by GLA gene, and to date over 1,000 mutations have been identified in the gene, most of which lead to synthesis of a non-functional protein [170, 171]. The incidence of Fabry disease is reported as 1 in 117,000 [172], however, the data may underestimate the exact disease frequency due to the rarity of the disease and undiagnosed patients [169, 170]. α-Gal A cleaves the terminal galactose from globotriaosylceramide (Gb3), deficiency of α-Gal A leads to progressive accumulation of Gb3 and its analogue lyso-Gb3 (globotriaosylsphingosine) within lysosomes in most cell types, thus affecting different tissues and organs [173,174]. Over time, lysosomal storage of these glycosphingolipids causes cellular dysfunction and damage, eventually leading to organ failure such as cardiovascular complications and end-stage renal disease [175, 176]. Although associated with side effects, enzyme replacement therapy (ERT) is the current standard of care for Fabry disease, with Replagal® and Fabrazyme® commercially available [177–179]. Another approach is chaperone therapy, as chaperones bind to defective α-Gal A and help with its functioning. However, a limitation of this therapy is that only the patients with amenable mutations can be treated [169]. Martini’s group reports the employment of mRNA coding α-Gal A towards treating Fabry disease. Notably, a single intravenous administration (0.5 mg/kg) of α-Gal A mRNA into Fabry mice achieved long-lasting (at least 12 weeks) attenuation of Gb3 accumulation in tissues, suggesting that less frequent dosing of mRNA therapy may be required compared to the conventional ERT therapy, which is administered biweekly [180].
Phenylketonuria (OMIM ID 261600)
Phenylketonuria (PKU), also known as phenylalanine hydroxylase (PAH) deficiency or Følling disease, is an autosomal recessive inborn error of phenylalanine (Phe) metabolism [181]. PKU is the most common metabolic defect of an amino acid in humans, with an average prevalence worldwide of ∼1:10,000 live births [182]. PAH is a hepatic enzyme responsible for converting Phe to tyrosine (Tyr) through Phe hydroxylation [181, 183]. PKU is caused predominantly by pathogenetic mutations in the PAH gene, and more than 1,000 such variants have been identified [184]. PAH deficiency results in accumulation of Phe in the body, leading to hyperphenylalaninaemia (HPA). Spontaneous deamination of Phe results in the formation of phenylpyruvate and other phenylketones, which are excreted in urine [182]. The toxic effect of HPA causes brain dysfunction, and if left untreated, PKU patients develop intellectual disability. Dietary restriction of Phe is the prevailing treatment, preventing intellectual disability in early treated patients. However, issues associated with medical foods include poor compliance (due to palatability of the diet) and nutritional deficiencies [185–187]. mRNA therapy is a promising alternative treatment for PKU. Notably, biweekly administration of PAH mRNA (1.0 mg/kg) encapsulated within lipid nanoparticles significantly reduced the level of serum Phe (from more than 1500 μM to less than 500 μM) in PAH knockout mice [188].
Pope disease (OMIM ID 232300)
Pompe disease (PD), the first documented LSD, is also known as glycogenosis type II, type II glycogen storage disease (GSDII), or acid maltase deficiency [189–192]. PD is inherited in an autosomal recessive manner, and is a chronic and severe metabolic myopathy caused by mutations in the gene encoding acid α-glucosidase (GAA) [189–191, 193, 194]. The estimated prevalence of PD, dependent on different ethnic groups and geographic regions, is ∼1–9 in 100,000 newborns [195–197]. In lysosomes, GAA degrades glycogen into glucoses in acidic pH by hydrolyzing glycogen’s α-1,4 and α-1,6-glucosidic linkages [198–200]. Partial or total GAA deficiency (due to mutations) induces lysosomal accumulation and storage of glycogen in cells of multiple tissues, leading to lysosomal rupture, cell malfunctions and autophagy [190, 192, 201]. Cardiac and skeletal muscles are the most affected tissues. Currently, ERT using recombinant human GAA (rhGAA) is the only clinically approved treatment for PD, improving the respiratory and cardiac functions of patients and increasing their life expectancy [202]. However, immunologic reactions against rhGAA and poor delivery and uptake of the enzyme to muscles limit the efficacy of ERT [191, 203]. An mRNA therapy for PD has been patented [204], providing a GAA mRNA encapsulated in lipid nanoparticles that can restore efficient and sustained enzyme expression in vivo, thus reducing the storage of glycogen in liver and muscle cells.
mRNA based cellular modulation
mRNA based cellular modulation is used in regenerative therapies to supplement defective tissue with molecular compounds necessary for restoring them to their functional physiological state. This restoration process requires a variety of proteins like growth factors, cytokines, and transcription factors that are essential for cell division and growth. You et al. successfully produced EVs by cellular nanoporation of human dermal fibroblasts and encapsulation of exogenous mRNA encoded with extracellular-matrix α1 type-I collagen (COL1A1). The EVs loaded with COL1A1 mRNA was intradermally delivered to mice with collagen depleted dermal tissue via microneedle arrays. The corresponding COL1A1 mRNA protein was identified starting from 12 h post and peaked at 4 days after delivery, successfully reducing the wrinkles in photoaged skin [111]. It is evident that mRNA can be potentially encoded with proteins that could modulate the cellular characterises. Yamanaka et al. designed mRNA encoding transcription factors such as OCT3/4, SOX2, MYC and KLF4 (currently known as Yamanaka factors). Transfecting these transcription factor-encoded mRNAs to somatic cells reprogramed them into induced pluripotent stem cells (iPSC) [275].
Sun et al. investigated the potential of using modified mRNA (AZD8601) encoded with vascular endothelial growth factor A (VEGF-A) as a therapeutic intervention for diabetic wound healing. The study displayed accelerated dose dependent neo-vessel formation, re-epithelialization and blood flow upregulation, facilitating tissue regeneration [276]. Above results were confirmed by the study conducted by Zha et al. where they delivered VEGF-A mRNA via LNP for wound healing in diabetic induced mice [277]. Recently, Antila et al. have been exploring the therapeutic potential of AZD8601 in regeneration of cardiac muscles after bypass surgery, however, they were unable to find a significant difference in efficacy in regeneration compared to the placebo in human trial even though promising results were observed in animal trials. Further studies are ongoing to improve the therapeutic efficacy and delivery [268, 269, 278].
mRNA-based immunomodulation
Basic human immunity can be divided into two main categories: active and passive immunity. This study reviews the utilization of synthetic mRNA to exploit the active and passive immune pathways in the human body as functional anti-viral and anti-cancer therapies (Figure 7). Pardi et al. developed the world’s first mRNA-based platform for delivering the heavy and light chains of the anti-HIV-1 antibody (VRC01) and demonstrated that mice transfected from mRNA encoded with anti-HIV-1 antibody (VRC01) produces VRCO1 antibodies [279, 280]. These findings highlight the feasibility of using encoded mRNA for both active and passive immune pathways for therapeutic intervention, laying the foundation of numerous innovations displayed below.
Anti-viral therapies
Human cytomegalovirus infections
Human cytomegalovirus (HHV-5) is a herpesvirus from the Herpesviridae family, transmitted via infected bodily fluids [281, 282]. Shwartz et al. conducted single-cell transcriptomic studies to elucidate the molecular pathogenesis in infected individuals involving HMCV latency and reactivation [281]. Previous studies identified glycoprotein subunit gB as a significant mediator of viral entry in fibroblast and epithelial cells [283, 284]. Epithelial and endothelial cells are often infected due to their expression of receptors that facilitate the entry of the pentameric complex comprising gH/gL/UL128-131 glycoproteins [285, 286].
Epidemiological data indicate that CMV poses a considerable global health challenge, affecting individuals of all ages [287]. Furthermore, among all live-born infants, the incidence of congenital cytomegalovirus (cCMV) infection is 1 in 200 in high-income countries and 1 in 71 in low-income countries [288].
Treating human cytomegalovirus (hCMV) is challenging, with FDA-approved drugs like cidofovir and ganciclovir contraindicated due to cross-resistance and toxicity. Current investigational hCMV vaccines, including mouse cytomegalovirus (mCMV) vaccine targeting viral G-protein-coupled receptors (M33), avoid systematic CMV spread only in animal models [285]. The gB/MF59 vaccine failed to block viral entry to epithelial cells [284]. Kschonsak (2022) revealed the structural basis for hCMV epitope engagement involving pentamer-thrombomodulin complex and viral entry mechanism [289]. Antibodies blocking viral entry have lower potency to neutralize glycoprotein B than those recognizing quarternary neutralising epitopes formed by pentameric complex-gH/gL/UL128/UL130/UL131 [284].
In 2023, Ciaramella (2023) (Moderna Inc., USA.) invented an mRNA-based LNP vaccine formulation (encoding pentameric complex +gB glycoprotein subunit plus compound 25: cholesterol) against hCMV [286]. This mRNA-based hCMV vaccine showed safe and effective immunogenic responses to prevent the maternal-fetal transmission of congenital viral diseases, especially herpes virus (HSV-5) [290]. The design rationale of this broad-spectrum mRNA vaccine against hCMV was to express at least one polypeptide encoding hCMV(gH, UL130, 131, UL 128) and one polypeptide expressing hCMV gB, both co-expressed within a non-viral delivery construct-a lipid-based nanoparticle formulation [286]. In Phase 1 trail, following both priming (4.2 µg) and boosting doses (1.4 µg) in in vivo models (balb/c mice model), this second generation hCMV mRNA vaccines encoding the pentamer and gB formulated with Compound 25 lipids elicited strong pentamer-specific antibody titres above 3 log10 at two different time points (day 40, 3 weeks post prime; day 40, 3 weeks post boost), measured by ELISA. Further experimental data highlighted the significance of using CytoGam (a hyperimmune serum for HCMV prophylaxis) as a benchmark to compare the neutralising HCMV antibody titers. Similarly, Moderna’s CMvictory phase 3 trial NCT05085366 is currently investigating safety, efficacy and immunogenicity of LNP-mRNA 1647 (multi-antigenic vaccine encoding pentameric variant: gL-UL128-UL130-UL131A and a glycoprotein complex) in healthy adults against CMV infection [291]. With robust long-lasting T and B cell responses, the Phase 3 trial was concluded with the promising therapeutic potential of mRNA-1647 vaccine and early prevention of CMV infection [286]. This implies that current investigational vaccines at various stages of clinical trial involving nano-engineered RNA technology and LNP formulations serve as an iconic prophylactic strategy. From ongoing clinical studies, they are found to be effective in the prevention and spread of congenital cytomegalovirus infection in high-risk groups (from pregnant mothers to unborn fetuses, infant patients and Allogenic hematopoietic cell transplanted patients) [286, 292].
Betacoronavirus
Betacoronaviruses (Beta CoVs), from the Coronavirinae subfamily, are pathogenic respiratory viruses that have severely impacted global populations [293, 294]. They are (30 kb) positive-sense, enveloped, single-stranded ribonucleic acid (RNA) viruses further comprising 4 lineages (A, B, C and D) [293]. SARS-CoV-2, an emerging Beta CoV, raised significant health and economic concerns [293, 294]. Beta lineage has affected over 90 countries [295]. Unlike other Beta CoVs such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), patients with COVID-19 are mostly asymptomatic [296]. Yet SARS-COV-2 infections are more life-threatening due to high contagiousness [294, 297]. Human-human transmission drives elevated cases, with bats and pangolins as potential intermediate vectors [293, 294].
Females are less susceptible to Beta CoVs due to higher biallelic X-linked immune-related gene expressions involving TLR-7 and cytotoxic T cell responses [293]. Sharma and colleagues have further shown that angiotensin-converting enzyme 2 (ACE 2) – a key viral contact and entry point for spike glycoproteins of SARS-CoV, is highly expressed in testis compared to ovaries [298]. During infection, these viruses elevate interleukins (IL-6 and IL-8) suggesting estrogen plays a stimulatory role in the sex-specific innate and adaptive immune responses [293].
Given the sex-specific differences in immunological reactions to both vaccination and COVID-19 infection, several sex hormonal therapies entered pre-clinical and clinical studies to treat both men and women using specific medical interventions [293, 299], for example, anti-androgen treatment (Clinical ID: NCT04446429) and progesterone for the treatment of COVID-19 in hospitalized men (Clinical ID: NCT04365127), both completed in 2021 [293]. Since the outbreak, various biomedical and pharmacotherapies entered the pipeline, among them are COVID-19 antiviral drugs of various classifications; such as remdesivir (nucleotide analogue), tocilizumab (biologics), various RNA synthesis inhibitors, and protease inhibitors like ritonavir/lopinavir combination therapies, etc [293, 299].
Understanding molecular pathogenesis and sex-specific immune responses has led to lipid-based nanoformulations with mRNA sequences against betacoronaviruses [294]. In 2020, Ciaramella and Himansu (Moderna Inc.) developed recombinant mRNA vaccines containing ORF encoding Spike glycoprotein [294]. This patented mRNA vaccine comprises of 2 constructs First, mRNA encoding coronavirus spike glycoprotein receptor binding domains. Second mRNA construct encoding proinflammatory cytokines (Interleukins, TGFs, interferons, TNF-α, M-CSFs, and GM-CSF) [294].
Today, only two mRNA-based vaccines against beta CoVs have received full US Food and Drug (FDA) approval, notably the Pfizer-BioNTech and Moderna COVID-19 vaccine [293]. By 2021, the unprecedented 5.42 million deaths due to COVID-19 and the red alert established by the WHO has reinforced the urgency of establishing an effective and safe non-viral RNA delivery platform [300, 301].
Lassa virus mRNA vaccine
The Lassa mammarena virus (LASV) from Arenaviridae is one of the most lethal viral haemorrhagic fever (VHF) viruses. VHF viruses can invade vascular organs causing cardiovascular dysfunction and hepatitis [302] LASV infection is endemic in Western Africa with 5,000 deaths yearly, with cases reported in the UK, Netherlands and Germany [302, 303]. Due to increasing seroprevalence and LASV lineages (I-VII), they are on WHO’s high-priority list [304].
LASV has an 80–200 nm enveloped RNA genome encoding four proteins: nucleoprotein (NP), glycoproteins complex (GPC), zinc-binding domain and large proteins. GPC enables viral attachment and entry through endosomal pathways [302]. A glycan shield around GPC helps evade host immune responses through epitope switching, blocking LASV-specific antibody neutralization [302, 305].
Currently, to treat fatal Lassa fever, many small molecules have entered clinical trials, such as favipiravir and ribavirin combination therapy and a novel LHF-535 (the enhanced analogue of benzimidazole derivative), which is a viral entry blocker [302, 306]. While other immunotherapies such as Arevirumab-3 succeeded in providing complete immunity against fatal Lassa fever, this monoclonal antibody cocktail has only been tested in non-human primates and rendered ineffective against the evolving Lassa virus lineage [302]. By 2023, CureVac AG Tübingen developed a prophylactic treatment for life-threatening Lassa fever using a non-viral LNP vector system [307]. Jasny (2023) created an mRNA-based vaccine encoding antigenic peptides expressing Lassa virus GPC and NP peptide, showing efficient responses at low doses. These inventors further demonstrated how mRNA-based vaccines can be engineered to encode different antigenic peptides expressing Lassa virus GPC precursor and NP peptide (from the highly evolving Lassa virus of clade I), showing efficient antigen-specific responses even at low dose regimen [307]. The therapeutic efficacy of the invented LASV-specific mRNA vaccine carrying a combination of antigenic peptides was evaluated in HeLa cells and mice models (balb/c) and induced robust T-cell and memory B-cell responses, with minimal side effects [307]. The current invention holds many promises and has now progressed to a phase I clinical trial. In a cohort study, the safety and efficacy profile of the LASV-specific mRNA vaccine was evaluated in human subjects following an intramuscular injection, receiving a prime-boost of 2 doses to complete the vaccination [307].
mRNA based anti-cancer vaccines
Cancer vaccines include preventative vaccines to impede cancer development in healthy individuals and therapeutic vaccines to treat existing cancer by enhancing immune responses. RNA vaccines, particularly mRNA polynucleotides, have superior design features for protein translation by utilizing existing cellular machinery. Unlike conventional vaccines produced externally, RNA vaccines are administered in a manner resembling the body’s natural state. Valiante et al., patented a cancer vaccine using mRNA that encodes multiple cancer antigens and an immune checkpoint modulator. The vaccine involves administering RNA polynucleotides containing an ORF encoding antigenic polypeptides, inducing specific immune responses comparable to vaccination [308].
Lung cancer therapies
Genomic alterations in lung cancer affect both oncogenes and tumour suppressor genes. Chromosomal rearrangements, mitotic recombination, point mutations, and epigenetic events like promoter methylation can inactivate tumour suppressor genes [309]. Key tumour suppressor genes in lung malignancies include CDKN2 (p16INK4a or MST1, 9p21), RB1 (13q14.11), TP53 (17p13.1), and several genes at 3p [310].
Kallen et al. developed a cancer vaccine containing six mRNAs with histone stem loops in 3′UTR regions, encoding different antigen [311]. These antigens - MAGE-C2, 5T4 (TPBG), MAGE-Ci, MUC1, NY-ESO-1 (CTAG1 B), and survivin (BIRC5) - induce adaptive immune responses in mammals. The vaccine includes at least one mRNA encoding antigen combinations and is designed to stimulate immune response for treating lung cancer, specifically NSCLC, and related disorders [311].
KRAS pathway-based cancer vaccines
Ashburn et al. designed an LNP-based mRNA cancer vaccine for use in human patients. The vaccine delivers two separate mRNAs: the first encodes a collection of peptide epitopes, while the second encodes epitopes specifically derived from common cancer hotspot mutations, such as KRAS G12 or G13. By incorporating both personalized antigens and shared cancer mutations, the approach aims to generate a stronger and broader immune response. This strategy is further integrated with anti-cancer immunotherapy to improve treatment outcomes [312].
Valiante et al. introduced concatemeric, or poly-epitope, mRNA cancer vaccines, in which a single mRNA construct encodes multiple cancer epitopes [313]. These vaccines are designed to target key mutations such as KRAS and p53 and can also incorporate immune stimulators or adjuvants like STING to boost immune responses. Packaged in LNPs, the vaccine may contain one or more mRNA molecules, each with an ORF encoding at least two peptide epitopes. The process begins with analyzing tumour biopsies to identify cancer-specific antigens and selecting immunogenic epitopes for inclusion in the mRNA construct. Once delivered intradermally, intramuscularly, or subcutaneously, the vaccine instructs the host’s cells to generate a wide spectrum of tumour-derived proteins or fragments, thereby promoting a strong and targeted anti-cancer immune response [313].
Personalized cancer vaccine
Zhong et al. revolutionized strategies for producing, administering vaccines, and formulating vaccine compositions [314]. The formulation of certain cancer vaccines incorporates a single mRNA with an open reading frame (ORF) that encodes fifteen peptide epitopes. This vaccine has the potential to facilitate the production of nearly any desired cancer protein or its segment through the body’s cellular mechanisms. The proposed protocol for treating a cancer patient involves analyzing a patient-derived sample to identify one or more personalized cancer antigens, which are then assessed for the anti-tumour efficacy of at least two peptide epitopes for each identified personalized cancer antigen. Once the epitopes have been identified, the cancer vaccine is developed [314].
Chen et al. developed a method to predict cancer patient response to MDM2 agonists (formulas I–III) by measuring mRNA expression of MDM2, along with a panel of genes such as CDKN2A, XPC, and BBC3 [315]. This biomarker-based approach helps identify patients likely to benefit from therapies targeting the MDM2–p53 interaction. The study assessed several biomarkers, including CDKN2A, TNFRSFOB, BBC3, CCNG1, TRIAP1, CDKN1A, MDM2, FDXR, DDB2, XPC, EDA2R, RPS27L, and BAX, comparing MDM2 mRNA expression in patients to reference levels from others with the same cancer type [315]. To support this, the authors used 281 human cancer cell lines (CELLO) derived from diverse tumour types and tissues. Genomic profiling revealed that 210 of these cell lines carried TP53 mutations, underscoring the importance of integrating biomarker analysis with targeted treatment strategies [315].
Prostate cancer
Prostate cells are vulnerable to mutations that promote uncontrolled proliferation, ultimately leading to prostate cancer. Several tumour-associated antigens—including Ep-CAM, HER-2, PSCA, PAP, PSMA, and PSA—are significantly overexpressed in prostate cancer cells compared to normal tissue, making them valuable therapeutic targets. Conventional treatments such as chemotherapy, hormone therapy, radiation, and proton therapy, whether used alone or in combination, often provide only limited benefit.
To overcome these challenges, Kallen et al. explored the use of mRNA-based vaccines designed to encode prostate cancer–associated antigens [316]. Key targets included PAP, MUC1, STEAP, PSA, PSMA, and PSCA, with the mRNAs administered either as a single formulation or as multiple constructs, each encoding one antigen. By inducing a strong adaptive immune response, this strategy aimed to enhance tumour recognition and elimination. The approach showed potential applicability across diverse prostate cancer subtypes, including adenocarcinoma, locally advanced disease, castration-resistant forms, and metastatic cancers [316].
Immune enhancer mRNA therapy
Huang et al. extracted mRNAs encoding polypeptides that enhance immune responses to specific antigens [317]. These antigens activate pathways like Type I interferon or NF-κB, and influence dendritic cell development and inflammatory responses. The mRNAs contain modified nucleobases and can augment immune reactions when co-administered with antigens against cancer or pathogens [317].
Frederick et al. developed therapeutic strategies for cancer management. mRNAs coding for cell-associated cytokines (IL-15, IL-12) and costimulatory molecules (OX40L) activate T cells and NK cells, showing anti-tumour efficacy in solid tumours and myeloid malignancies [318]. The combination of mRNAs encoding IL-15, IL-12, and OX40L enhances anti-tumour treatment by efficiently activating T cells and NK cells [318].
Frederick et al. also proposed anti-cancer mRNA-combined therapies, combining at least two combined polynucleotides (mRNAs) [318]. The polynucleotides are limited to: (i) a polynucleotide encoding IL23 as immune response primer; (ii) a polynucleotide encoding OX40L as immune response co-stimulatory signal; (iii) a polynucleotide encoding anti-CTLA-4 antibody as checkpoint inhibitor; or (iv) their combinations. The therapy aims to reduce tumor size or inhibit growth in patients through intra-tumoural injection of these combinations [319].
Future perspectives in mRNA therapeutics
The landscape of mRNA therapeutics is evolving rapidly, and several directions appear particularly important for the years ahead. One area attracting considerable attention is the development of cancer vaccines. Instead of applying a “one-size-fits-all” approach, future strategies are likely to rely on tailoring vaccines to the mutational signatures of individual tumours. Such personalization has the potential to improve therapeutic benefit while limiting unwanted side effects, making vaccines a central component of precision oncology.
Another avenue of progress lies in the treatment of inherited metabolic and genetic disorders. Early work in conditions such as citrullinemia type II demonstrates that mRNA can act as a temporary source of missing proteins. With refinement, the same principle could be extended to a much wider group of disorders where protein replacement is a rational therapy. At the same time, cancer research is exploring how mRNA treatments might be combined with existing modalities—checkpoint inhibitors, adoptive cell therapies, or even conventional chemotherapy—to produce stronger and longer-lasting responses than any single treatment alone.
Underlying all of these possibilities is the critical issue of delivery. mRNA is inherently unstable and easily degraded, so the development of more sophisticated carriers remains a central challenge. Advances in lipid nanoparticles, polymer formulations, and naturally derived vesicles are beginning to address these hurdles, and continued progress here will be essential if mRNA therapies are to reach their full range of applications. Importantly, these innovations are opening opportunities beyond oncology, with infectious disease prevention, neurodegeneration, cardiovascular disease, and autoimmune disorders all emerging as promising targets.
Scientific progress must, however, be matched with social responsibility. Questions of equitable access, transparent regulation, and long-term safety monitoring will be decisive in shaping the public’s trust. Education of both healthcare professionals and patients will also be vital to ensure that the benefits of mRNA technology are widely understood and accepted.
Finally, no single group can achieve these advances in isolation. The field will benefit most from strong collaborations linking academic laboratories, clinicians, industry partners, and regulatory authorities. Such partnerships will accelerate discovery and help to ensure that effective therapies move swiftly from the bench to the bedside.
In conclusion, mRNA therapeutics represent a genuine shift in modern medicine. Continued scientific innovation, coupled with thoughtful attention to delivery, accessibility, and collaboration, holds the promise of transforming care across a wide spectrum of diseases.
Statements
Author contributions
The collaborative effort behind this study involved multiple researchers with distinct roles and contributions. SH took the lead in conceptualizing the study, creating the manuscript’s structure, gathering data, and producing the initial draft. SC and SB played equal parts in writing the original draft and designing the figures, with SB providing additional input on mRNA encapsulation and delivery. The team’s expertise was further utilized as SC, QZ, and KR focused on drafting sections related to mRNA-based therapeutic applications and managing the citation process. RV offered supervision and resources, while also joining BP and Steve SW in reviewing and editing the original draft. This collaborative approach culminated in a final review and approval process involving all listed authors, who collectively ensured the manuscript’s quality, transparency, and accountability. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors express their gratitude for the invaluable support and resources provided by the Perron Institute and the Murdoch University Personalised Medicine Institute. SH and SB are particularly thankful for the funding from the Perron Institute for Neurological and Translational Science and the International Tuition Fee Scholarship (ITFS) from Murdoch University, which played a significant role in advancing this work.
Conflict of interest
SW is a consultant to Sarepta Therapeutics and is an inventor on patents related to exon-skipping therapies for Duchenne muscular dystrophy. These patents are licensed to Sarepta Therapeutics and may be relevant to the technologies discussed in this manuscript. RV is a Co-Founder, Director Board Member of ProGenis Pharmaceuticals, a company engaged in the development of RNA-based therapeutics. He is also an inventor on multiple patent applications regarding oligonucleotide synthesis and therapeutic development. These affiliations and intellectual property interests have been disclosed and are managed in accordance with institutional and journal policies.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
References
1.
KulkarniJAWitzigmannDThomsonSBChenSLeavittBRCullisPRet alThe current landscape of nucleic acid therapeutics. Nat Nanotechnology (2021) 16(6):630–43. 10.1038/s41565-021-00898-0
2.
SparmannAVogelJ. RNA-based medicine: from molecular mechanisms to therapy. The EMBO J (2023) 42(21):e114760. 10.15252/embj.2023114760
3.
OuranidisAVavilisTMandalaEDavidopoulouCStamoulaEMarkopoulouCKet almRNA therapeutic modalities design, formulation and manufacturing under pharma 4.0 principles. Biomedicines (2021) 10(1):50. 10.3390/biomedicines10010050
4.
DjebaliSDavisCAMerkelADobinALassmannTMortazaviAet alLandscape of transcription in human cells. Nature (2012) 489(7414):101–8. 10.1038/nature11233
5.
DaiXZhangSZaleta-RiveraK. RNA: interactions drive functionalities. Mol Biol Rep (2020) 47(2):1413–34. 10.1007/s11033-019-05230-7
6.
KarikóKMuramatsuHWelshFALudwigJKatoHAkiraSet alIncorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther (2008) 16(11):1833–40. 10.1038/mt.2008.200
7.
IbbaMLCicconeGEspositoCLCatuognoSGiangrandePH. Advances in mRNA non-viral delivery approaches. Adv Drug Deliv Rev (2021) 177:113930. 10.1016/j.addr.2021.113930
8.
WolffJAMaloneRWWilliamsPChongWAcsadiGJaniAet alDirect gene transfer into mouse muscle in vivo. Science (1990) 247(4949):1465–8. 10.1126/science.1690918
9.
MartinonFKrishnanSLenzenGMagnéRGomardEGuilletJ-Get alInduction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur J Immunol (1993) 23(7):1719–22. 10.1002/eji.1830230749
10.
HouXZaksTLangerRDongY. Lipid nanoparticles for mRNA delivery. Nat Rev Mater (2021) 6(12):1078–94. 10.1038/s41578-021-00358-0
11.
LiuAWangX. The pivotal role of chemical modifications in mRNA therapeutics. Front Cell Dev Biol (2022) 10:901510. 10.3389/fcell.2022.901510
12.
VavilisTStamoulaEAinatzoglouASachinidisALamprinouMDardalasIet almRNA in the context of protein replacement therapy. Pharmaceutics (2023) 15(1):166. 10.3390/pharmaceutics15010166
13.
JohnsonR. Medicine from modified mRNA. Nat Chem Biol (2023) 19(12):1443. 10.1038/s41589-023-01498-w
14.
AndersonBRMuramatsuHNallagatlaSRBevilacquaPCSansingLHWeissmanDet alIncorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res (2010) 38(17):5884–92. 10.1093/nar/gkq347
15.
RitchieHMathieuERodés-GuiraoLAppelCGiattinoCOrtiz-OspinaEet alCoronavirus pandemic (COVID-19): ourworldindata.org (2020). Available online at: https://ourworldindata.org/covid-vaccinations#citation.
16.
FrancoMKKoutmouKS. Chemical modifications to mRNA nucleobases impact translation elongation and termination. Biophysical Chem (2022) 285:106780. 10.1016/j.bpc.2022.106780
17.
BaptistaBCarapitoRLarouiNPichonCSousaF. mRNA, a revolution in biomedicine. Pharmaceutics (2021) 13(12):2090. 10.3390/pharmaceutics13122090
18.
KimSCSekhonSSShinWRAhnGChoBKAhnJYet alModifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Mol Cell Toxicol (2022) 18(1):1–8. 10.1007/s13273-021-00171-4
19.
HouXShiJXiaoY. mRNA medicine: recent progresses in chemical modification, design, and engineering. Nano Res (2024) 17(10):9015–30. 10.1007/s12274-024-6978-6
20.
KarikóKBucksteinMNiHWeissmanD. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity (2005) 23(2):165–75. 10.1016/j.immuni.2005.06.008
21.
KarikóKWeissmanD. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr Opin Drug Discov Devel (2007) 10(5):523–32. 10.1016/j.immuni.2005.06.008
22.
AndriesOMc CaffertySDe SmedtSCWeissRSandersNNKitadaT. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Controlled Release (2015) 217:337–44. 10.1016/j.jconrel.2015.08.051
23.
MoraisPAdachiHYuYT. The critical contribution of pseudouridine to mRNA COVID-19 vaccines. Front Cell Dev Biol (2021) 9:789427. 10.3389/fcell.2021.789427
24.
BéroutiMWagnerMGreulichWPisedduIGärtigJHansbauerLet alPseudouridine RNA avoids immune detection through impaired endolysosomal processing and TLR engagement. Cell (2025) 188(18):4880–95.e15. 10.1016/j.cell.2025.05.032
25.
MulroneyTEPöyryTYam-PucJCRustMHarveyRFKalmarLet alN1-methylpseudouridylation of mRNA causes +1 ribosomal frameshifting. Nature (2024) 625(7993):189–94. 10.1038/s41586-023-06800-3
26.
KomarAA. A code within a code: how codons fine-tune protein folding in the cell. Biochemistry (Mosc) (2021) 86(8):976–91. 10.1134/s0006297921080083
27.
MauroVP. Codon optimization in the production of recombinant biotherapeutics: potential risks and considerations. BioDrugs (2018) 32(1):69–81. 10.1007/s40259-018-0261-x
28.
HansonGCollerJ. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol (2018) 19(1):20–30. 10.1038/nrm.2017.91
29.
DingWYuWChenYLaoLFangYFangCet alRare codon recoding for efficient noncanonical amino acid incorporation in mammalian cells. Science (2024) 384(6700):1134–42. 10.1126/science.adm8143
30.
PresnyakVAlhusainiNChenYHMartinSMorrisNKlineNet alCodon optimality is a major determinant of mRNA stability. Cell (2015) 160(6):1111–24. 10.1016/j.cell.2015.02.029
31.
SaikiaMWangXMaoYWanJPanTQianSB. Codon optimality controls differential mRNA translation during amino acid starvation. Rna (2016) 22(11):1719–27. 10.1261/rna.058180.116
32.
ParmleyJLHuynenMA. Clustering of codons with rare cognate tRNAs in human genes suggests an extra level of expression regulation. PLoS Genet (2009) 5:e1000548. 10.1371/journal.pgen.1000548
33.
WangYLiCKhanMRWangYRuanYZhaoBet alAn engineered rare codon device for optimization of Metabolic pathways. Sci Rep (2016) 6:20608. 10.1038/srep20608
34.
ZhangHZhangLLinAXuCLiZLiuKet alAlgorithm for optimized mRNA design improves stability and immunogenicity. Nature (2023) 621(7978):396–403. 10.1038/s41586-023-06127-z
35.
DiezMMedina-MuñozSGCastellanoLAda Silva PescadorGWuQBazziniAA. iCodon customizes gene expression based on the codon composition. Scientific Rep (2022) 12(1):12126. 10.1038/s41598-022-15526-7
36.
WuXShanKJZanFTangXQianZLuJ. Optimization and deoptimization of codons in SARS-CoV-2 and related implications for vaccine development. Adv Sci (2023) 10(23):e2205445. 10.1002/advs.202205445
37.
WalczakSNowickaAKubackaDFacKWanatPMroczekSet alA novel route for preparing 5' cap mimics and capped RNAs: phosphate-modified cap analogues obtained via click chemistry. Chem Sci (2017) 8(1):260–7. 10.1039/c6sc02437h
38.
SperlingS. Transcriptional regulation at a glance. BMC Bioinformatics (2007) 8(6):S2. 10.1186/1471-2105-8-s6-s2
39.
RamanathanARobbGBChanSH. mRNA capping: biological functions and applications. Nucleic Acids Res (2016) 44(16):7511–26. 10.1093/nar/gkw551
40.
TopisirovicISvitkinYVSonenbergNShatkinAJ. Cap and cap-binding proteins in the control of gene expression. WIREs RNA (2011) 2(2):277–98. 10.1002/wrna.52
41.
FechterPBrownleeGG. Recognition of mRNA cap structures by viral and cellular proteins. J Gen Virol (2005) 86(Pt 5):1239–49. 10.1099/vir.0.80755-0
42.
DespicVJaffreySR. mRNA ageing shapes the Cap2 methylome in mammalian mRNA. Nature (2023) 614(7947):358–66. 10.1038/s41586-022-05668-z
43.
WarminskiMDepaixAZiemkiewiczKSpiewlaTZuberekJDrazkowskaKet alTrinucleotide cap analogs with triphosphate chain modifications: synthesis, properties, and evaluation as mRNA capping reagents. Nucleic Acids Res (2024) 52(18):10788–809. 10.1093/nar/gkae763
44.
MuttachFMuthmannNRentmeisterA. Synthetic mRNA capping. Beilstein J Org Chem (2017) 13:2819–32. 10.3762/bjoc.13.274
45.
HendersonJMUjitaAHillEYousif‐RosalesSSmithCKoNet alCap 1 messenger RNA synthesis with Co-transcriptional CleanCap® analog by in vitro transcription. Curr Protoc (2021) 1(2):e39. 10.1002/cpz1.39
46.
GrzelaRPiecykKStankiewicz-DrogonAPietrowPLukaszewiczMKurpiejewskiKet alN2 modified dinucleotide cap analogs as a potent tool for mRNA engineering. Rna (2023) 29(2):200–16. 10.1261/rna.079460.122
47.
ShanmugasundaramMSenthilvelanAKoreAR. Recent advances in modified cap analogs: synthesis, biochemical properties, and mRNA based vaccines. The Chem Rec (2022) 22(8):e202200005. 10.1002/tcr.202200005
48.
OhnoHAkamineSMochizukiMHayashiKAkichikaSSuzukiTet alVersatile strategy using vaccinia virus-capping enzyme to synthesize functional 5' cap-modified mRNAs. Nucleic Acids Res (2023) 51(6):e34. 10.1093/nar/gkad019
49.
SenthilvelanAVonderfechtTShanmugasundaramMPalIPotterJKoreAR. Trinucleotide cap analogue bearing a locked nucleic acid moiety: synthesis, mRNA modification, and translation for therapeutic applications. Org Lett (2021) 23(11):4133–6. 10.1021/acs.orglett.1c01037
50.
KlöckerNWeissenboeckFPvan DülmenMŠpačekPHüwelSRentmeisterA. Photocaged 5′ cap analogues for optical control of mRNA translation in cells. Nat Chem (2022) 14(8):905–13. 10.1038/s41557-022-00972-7
51.
TarbashevichKGhoshADasAKuilyaDSharmaSNSinhaSet alOptochemical control over mRNA translation by photocaged phosphorodiamidate morpholino oligonucleotides in vivo. Nat Commun (2025) 16(1):3614. 10.1038/s41467-025-58207-5
52.
WilkieGSDicksonKSGrayNK. Regulation of mRNA translation by 5'- and 3'-UTR-binding factors. Trends Biochem Sci (2003) 28(4):182–8. 10.1016/s0968-0004(03)00051-3
53.
LeppekKDasRBarnaM. Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol (2018) 19(3):158–74. 10.1038/nrm.2017.103
54.
SonenbergNHinnebuschAG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell (2009) 136(4):731–45. 10.1016/j.cell.2009.01.042
55.
SzostakEGebauerF. Translational control by 3'-UTR-binding proteins. Brief Funct Genomics (2013) 12(1):58–65. 10.1093/bfgp/els056
56.
MayrC. What are 3' UTRs doing?Cold Spring Harb Perspect Biol (2019) 11(10):a034728. 10.1101/cshperspect.a034728
57.
AsraniKHFarelliJDStahleyMRMillerRLChengCJSubramanianRRet alOptimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol (2018) 15(6):756–62. 10.1080/15476286.2018.1450054
58.
Linares-FernándezSMorenoJLambertEMercier-GouyPVachezLVerrierBet alCombining an optimized mRNA template with a double purification process allows strong expression of in vitro transcribed mRNA. Mol Ther - Nucleic Acids (2021) 26:945–56. 10.1016/j.omtn.2021.10.007
59.
Castillo-HairSFedakSWangBLinderJHavensKCertoMet alOptimizing 5’UTRs for mRNA-delivered gene editing using deep learning. Nat Commun (2024) 15(1):5284. 10.1038/s41467-024-49508-2
60.
CaoJNovoaEMZhangZChenWCWLiuDChoiGCGet alHigh-throughput 5′ UTR engineering for enhanced protein production in non-viral gene therapies. Nat Commun (2021) 12(1):4138. 10.1038/s41467-021-24436-7
61.
PengJMurrayELSchoenbergDR. In vivo and in vitro analysis of poly(A) length effects on mRNA translation. Methods In Mol Biology™ (2008) 419:215–30. 10.1007/978-1-59745-033-1_15
62.
ElangoNElangoSShivshankarPKatzMS. Optimized transfection of mRNA transcribed from a d(A/T)100 tail-containing vector. Biochem Biophysical Res Commun (2005) 330(3):958–66. 10.1016/j.bbrc.2005.03.067
63.
JalkanenALColemanSJWiluszJ. Determinants and implications of mRNA poly(A) tail size--does this protein make my tail look big?Semin Cell & Developmental Biol (2014) 34:24–32. 10.1016/j.semcdb.2014.05.018
64.
PassmoreLACollerJ. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat Rev Mol Cell Biol (2022) 23(2):93–106. 10.1038/s41580-021-00417-y
65.
HoltkampSKreiterSSelmiASimonPKoslowskiMHuberCet alModification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood (2006) 108(13):4009–17. 10.1182/blood-2006-04-015024
66.
OhSKesslerJA. Design, assembly, production, and transfection of synthetic modified mRNA. Methods. (2018) 133:29–43. 10.1016/j.ymeth.2017.10.008
67.
Avci-AdaliMBehringASteinleHKellerTKrajeweskiSSchlensakCet alIn vitro synthesis of modified mRNA for induction of protein expression in human cells. J visualized experiments : JoVE (2014)(93) e51943–e. 10.3791/51943
68.
PreskeyDAllisonTFJonesMMamchaouiKUngerC. Synthetically modified mRNA for efficient and fast human iPS cell generation and direct transdifferentiation to myoblasts. Biochem Biophysical Res Commun (2016) 473(3):743–51. 10.1016/j.bbrc.2015.09.102
69.
GrandiCEmmaneelMNelissenFHTRoosenboomLWMPetrovaYElzoklaOet alDecoupled degradation and translation enables noise modulation by poly(A) tails. Cell Syst (2024) 15(6):526–43.e7. 10.1016/j.cels.2024.05.004
70.
HeWZhangXZouYLiJWangCHeYet alEffective synthesis of high-integrity mRNA using in vitro transcription. Molecules (2024) 29(11):2461. 10.3390/molecules29112461
71.
WebbCIpSBathulaNVPopovaPSorianoSKVLyHHet alCurrent status and future perspectives on MRNA drug manufacturing. Mol Pharmaceutics (2022) 19(4):1047–58. 10.1021/acs.molpharmaceut.2c00010
72.
AbeHKimuraYHonmaMNakamotoKMotosawaKAtagoTet alPosition-specific nucleoside sugar modifications in mRNA ORF: enhancing translational function through complete chemical synthesis of mRNA (2024).
73.
AbeNImaedaAInagakiMLiZKawaguchiDOndaKet alComplete chemical synthesis of minimal messenger RNA by efficient chemical capping reaction. ACS Chem Biol (2022) 17(6):1308–14. 10.1021/acschembio.1c00996
74.
HusseiniRAAbeNHaraTAbeHKogureK. Use of iontophoresis technology for transdermal delivery of a minimal mRNA vaccine as a potential melanoma therapeutic. Biol Pharm Bull (2023) 46(2):301–8. 10.1248/bpb.b22-00746
75.
CurryEMuirGQuJKisZHulleyMBrownA. Engineering an Escherichia coli based in vivo mRNA manufacturing platform. Biotechnol Bioeng (2024) 121(6):1912–26. 10.1002/bit.28684
76.
RyczekMPlutaMBłaszczykLKiliszekA. Overview of methods for large-scale RNA synthesis. Appl Sci (2022) 12(3):1543. 10.3390/app12031543
77.
KangDDLiHDongY. Advancements of in vitro transcribed mRNA (IVT mRNA) to enable translation into the clinics. Adv Drug Deliv Rev (2023) 199:114961. 10.1016/j.addr.2023.114961
78.
KarikóKMuramatsuHLudwigJWeissmanD. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res (2011) 39(21):e142. 10.1093/nar/gkr695
79.
SchoenmakerLWitzigmannDKulkarniJAVerbekeRKerstenGJiskootWet almRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int J pharmaceutics (2021) 601:120586. 10.1016/j.ijpharm.2021.120586
80.
CaoYGaoGF. mRNA vaccines: a matter of delivery. EClinicalMedicine (2021) 32:100746. 10.1016/j.eclinm.2021.100746
81.
PaunovskaKLoughreyDDahlmanJE. Drug delivery systems for RNA therapeutics. Nat Rev Genet (2022) 23(5):265–80. 10.1038/s41576-021-00439-4
82.
DowdySF. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol (2017) 35(3):222–9. 10.1038/nbt.3802
83.
VanderBurghJACorsoTNLevySLCraigheadHG. Scalable continuous-flow electroporation platform enabling T cell transfection for cellular therapy manufacturing. Scientific Rep (2023) 13(1):6857. 10.1038/s41598-023-33941-2
84.
QiuPZiegelhofferPSunJYangN. Gene gun delivery of mRNA in situ results in efficient transgene expression and genetic immunization. Gene Ther (1996) 3(3):262–8.
85.
MoodySA. Microinjection of mRNAs and oligonucleotides. Cold Spring Harb Protoc (2018) 2018(12):pdb.prot097261. 10.1101/pdb.prot097261
86.
Campillo-DavoDDe LaereMRoexGVerstevenMFlumensDBernemanZNet alThe ins and outs of messenger RNA electroporation for physical gene delivery in immune cell-based therapy. Pharmaceutics (2021) 13(3):396. 10.3390/pharmaceutics13030396
87.
RamachandranSSatapathySRDuttaT. Delivery strategies for mRNA vaccines. Pharm Med (2022) 36(1):11–20. 10.1007/s40290-021-00417-5
88.
Gómez-AguadoIRodríguez-CastejónJVicente-PascualMRodríguez-GascónASolinísMÁdel Pozo-RodríguezA. Nanomedicines to deliver mRNA: state of the art and future perspectives. Nanomaterials (2020) 10(2):364. 10.3390/nano10020364
89.
AfoninKADobrovolskaiaMAChurchGBatheM. Opportunities, barriers, and a strategy for overcoming translational challenges to therapeutic nucleic acid nanotechnology. Ther RNA Nanotechnology (2021) 605–23.
90.
SajidMIMoazzamMKatoSYeseom ChoKTiwariRK. Overcoming barriers for siRNA therapeutics: from bench to bedside. Pharmaceuticals (2020) 13(10):294. 10.3390/ph13100294
91.
BaiWYangDZhaoYLiGLiuZXiongPet alMulti-step engineered adeno-associated virus enables whole-brain mRNA delivery. bioRxiv. (2024). 2024.06.04.597261.
92.
ParkAHongPWonSTThibaultPAVigantFOguntuyoKYet alSendai virus, an RNA virus with no risk of genomic integration, delivers CRISPR/Cas9 for efficient gene editing. Mol Ther - Methods & Clin Development (2016) 3:16057. 10.1038/mtm.2016.57
93.
StuartASShawRHLiuXGreenlandMAleyPKAndrewsNJet alImmunogenicity, safety, and reactogenicity of heterologous COVID-19 primary vaccination incorporating mRNA, viral-vector, and protein-adjuvant vaccines in the UK (Com-COV2): a single-blind, randomised, phase 2, non-inferiority trial. The Lancet (2022) 399(10319):36–49. 10.1016/s0140-6736(21)02718-5
94.
ToudeshkchoueiMGTavakoliAMohammadghasemiHKarimiAAiJRabieeMet alRecent approaches to mRNA vaccine delivery by lipid-based vectors prepared by continuous-flow microfluidic devices. Future Med Chem (2022) 14(21):1561–81. 10.4155/fmc-2022-0027
95.
MendonçaMCPKontAKowalskiPSO'DriscollCM. Design of lipid-based nanoparticles for delivery of therapeutic nucleic acids. Drug Discov Today (2023) 28(3):103505. 10.1016/j.drudis.2023.103505
96.
GaoMTangMHoWTengYChenQBuLet alModulating plaque inflammation via targeted mRNA nanoparticles for the treatment of atherosclerosis. ACS nano (2023) 17(18):17721–39. 10.1021/acsnano.3c00958
97.
MartensTFRemautKDemeesterJDe SmedtSCBraeckmansK. Intracellular delivery of nanomaterials: how to catch endosomal escape in the act. Nano Today (2014) 9(3):344–64. 10.1016/j.nantod.2014.04.011
98.
ZukancicDSuysEJPilkingtonEHAlgarniAAl-WassitiHTruongNP. The importance of poly (ethylene glycol) and lipid structure in targeted gene delivery to lymph nodes by lipid nanoparticles. Pharmaceutics (2020) 12(11):1068. 10.3390/pharmaceutics12111068
99.
LiMFangEWangYShiLLiJPengQet alAn mRNA vaccine against rabies provides strong and durable protection in mice. Front Immunol (2023) 14:1288879. 10.3389/fimmu.2023.1288879
100.
LiJYuPLiuQXuLChenYLiYet alSafety and efficacy assessment of an mRNA rabies vaccine in dogs, rodents, and cynomolgus macaques. npj Vaccin (2024) 9(1):130. 10.1038/s41541-024-00925-w
101.
FangZYuPZhuW. Development of mRNA rabies vaccines. Hum Vaccin & Immunother (2024) 20(1):2382499. 10.1080/21645515.2024.2382499
102.
BaiSYangTZhuCFengMZhangLZhangZet alA single vaccination of nucleoside-modified rabies mRNA vaccine induces prolonged highly protective immune responses in mice. Front Immunol (2023) 13:1099991. 10.3389/fimmu.2022.1099991
103.
QinSTangXChenYChenKFanNXiaoWet almRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduction Targeted Therapy (2022) 7(1):166. 10.1038/s41392-022-01007-w
104.
YongHLinLLiZGuoRWangCLiuSet alTailoring highly branched poly (β-amino ester) s for efficient and Organ-selective mRNA delivery. Nano Lett (2024) 24(30):9368–76. 10.1021/acs.nanolett.4c02440
105.
ChandaPKSukhovershinRCookeJP. mRNA-enhanced cell therapy and cardiovascular regeneration. Cells (2021) 10(1):187. 10.3390/cells10010187
106.
KisakovaLAApartsinEKNizolenkoLFKarpenkoLI. Dendrimer-mediated delivery of DNA and RNA vaccines. Pharmaceutics (2023) 15(4):1106. 10.3390/pharmaceutics15041106
107.
AmiriABagherifarRAnsari DezfouliEKiaieSHJafariRRamezaniR. Exosomes as bio-inspired nanocarriers for RNA delivery: preparation and applications. J Translational Med (2022) 20(1):125. 10.1186/s12967-022-03325-7
108.
PegtelDMGouldSJ. Exosomes. Annu Rev Biochem (2019) 88:487–514. 10.1146/annurev-biochem-013118-111902
109.
TsaiS-JGuoCAtaiNAGouldSJ. Exosome-mediated mRNA delivery for SARS-CoV-2 vaccination. BioRxiv. (2020). 2020.11. 06.371419.
110.
YangTFogartyBLaForgeBAzizSPhamTLaiLet alDelivery of small interfering RNA to inhibit vascular endothelial growth factor in zebrafish using natural brain endothelia cell-secreted exosome nanovesicles for the treatment of brain cancer. The AAPS J (2017) 19:475–86. 10.1208/s12248-016-0015-y
111.
YouYTianYYangZShiJKwakKJTongYet alIntradermally delivered mRNA-encapsulating extracellular vesicles for collagen-replacement therapy. Nat Biomed Eng (2023) 7(7):887–900. 10.1038/s41551-022-00989-w
112.
PopowskiKDLópez de Juan AbadBGeorgeASilkstoneDBelcherEChungJet alInhalable exosomes outperform liposomes as mRNA and protein drug carriers to the lung. Extracellular Vesicle (2022) 1:100002. 10.1016/j.vesic.2022.100002
113.
TaylorDDShahS. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods. (2015) 87:3–10. 10.1016/j.ymeth.2015.02.019
114.
RosasLEElgamalOAMoXPhelpsMASchmittgenTDPapenfussTL. In vitro immunotoxicity assessment of culture-derived extracellular vesicles in human monocytes. J immunotoxicology (2016) 13(5):652–65. 10.3109/1547691x.2016.1148089
115.
ColaoILCortelingRBracewellDWallI. Manufacturing exosomes: a promising therapeutic platform. Trends Molecular Medicine (2018) 24(3):242–56. 10.1016/j.molmed.2018.01.006
116.
AslanCKiaieSHZolbaninNMLotfinejadPRamezaniRKashanchiFet alExosomes for mRNA delivery: a novel biotherapeutic strategy with hurdles and hope. BMC Biotechnol (2021) 21:20–12. 10.1186/s12896-021-00683-w
117.
DasA. Extracellular vesicles: tiny messengers for mighty RNA delivery. Biologics (2024) 4(1):88–104. 10.3390/biologics4010007
118.
FuXChenTSongYFengCChenHZhangQet almRNA delivery by a pH-Responsive DNA nano-hydrogel. Small (2021) 17(29):e2101224. 10.1002/smll.202101224
119.
ZhongRTalebianSMendesBBWallaceGLangerRCondeJet alHydrogels for RNA delivery. Nat Mater (2023) 22(7):818–31. 10.1038/s41563-023-01472-w
120.
YanJChenRZhangHBryersJD. Injectable biodegradable chitosan-alginate 3D porous gel scaffold for mRNA vaccine delivery. Macromolecular Biosci (2019) 19(2):e1800242. 10.1002/mabi.201800242
121.
RwandamuriyeFXVitaliBSchreursJWangTBarrickEIyerKSet alProtocol for delivery of intraoperative immunotherapy to mice by surgical debulking of subcutaneous tumors. STAR Protoc (2024) 5(2):102948. 10.1016/j.xpro.2024.102948
122.
RwandamuriyeFXEvansCWWylieBNorretMVitaliBHoDet alA surgically optimized intraoperative poly(I:C)-releasing hydrogel prevents cancer recurrence. Cell Rep Med (2023) 4(7):101113. 10.1016/j.xcrm.2023.101113
123.
SahinUKarikóKTüreciÖ. mRNA-based therapeutics — developing a new class of drugs. Nat Rev Drug Discov (2014) 13(10):759–80. 10.1038/nrd4278
124.
LyuMChenJPengYHanFGongLGuoJet alThe global patent landscape of mRNA for diagnosis and therapy. Nat Biotechnol (2023) 41(9):1193–9. 10.1038/s41587-023-01925-2
125.
PatelABahMAWeinerDB. In vivo delivery of nucleic acid-encoded monoclonal antibodies. BioDrugs (2020) 34(3):273–93. 10.1007/s40259-020-00412-3
126.
Van HoeckeLRooseK. How mRNA therapeutics are entering the monoclonal antibody field. J Transl Med (2019) 17(1):54. 10.1186/s12967-019-1804-8
127.
ParayathNNStephanSBKoehneALNelsonPSStephanMT. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat Commun (2020) 11(1):6080. 10.1038/s41467-020-19486-2
128.
XuSYangKLiRZhangL. mRNA vaccine Era—mechanisms, drug platform and clinical prospection. Int J Mol Sci (2020) 21(18):6582. 10.3390/ijms21186582
129.
KwonHKimMSeoYMoonYSLeeHJLeeKet alEmergence of synthetic mRNA: in vitro synthesis of mRNA and its applications in regenerative medicine. Biomaterials (2018) 156:172–93. 10.1016/j.biomaterials.2017.11.034
130.
Silvera-RuizSMArranzJAHäberleJAngaroniCJBezardMGuelbertNet alUrea cycle disorders in Argentine patients: clinical presentation, biochemical and genetic findings. Orphanet J Rare Dis (2019) 14(1):203–8. 10.1186/s13023-019-1177-3
131.
HayasakaK. Metabolic basis and treatment of citrin deficiency. J Inherit Metab Dis (2021) 44(1):110–7. 10.1002/jimd.12294
132.
OkanoYOhuraTSakamotoOInuiA. Current treatment for citrin deficiency during NICCD and adaptation/compensation stages: strategy to prevent CTLN2. Mol Genet Metab (2019) 127(3):175–83. 10.1016/j.ymgme.2019.06.004
133.
OkanoYOkamotoMYazakiMInuiAOhuraTMurayamaKet alAnalysis of daily energy, protein, fat, and carbohydrate intake in citrin-deficient patients: towards prevention of adult-onset type II citrullinemia. Mol Genet Metab (2021) 133(1):63–70. 10.1016/j.ymgme.2021.03.004
134.
CaoJAnDGaldurozMZhuoJLiangSEybyeMet almRNA therapy improves metabolic and behavioral abnormalities in a murine model of citrin deficiency. Mol Ther (2019) 27(7):1242–51. 10.1016/j.ymthe.2019.04.017
135.
KomatsuMTanakaNKimuraTYazakiM. Citrin deficiency: clinical and nutritional features. Nutrients (2023) 15(10):2284. 10.3390/nu15102284
136.
TavoulariSLacabanneDThangaratnarajahCKunjiER. Pathogenic variants of the mitochondrial aspartate/glutamate carrier causing citrin deficiency. Trends Endocrinol & Metab (2022) 33:539–53. 10.1016/j.tem.2022.05.002
137.
SahekiTKobayashiK. Mitochondrial aspartate glutamate carrier (citrin) deficiency as the cause of adult-onset type II citrullinemia (CTLN2) and idiopathic neonatal hepatitis (NICCD). J Hum Genet (2002) 47(7):333–41. 10.1007/s100380200046
138.
BadmusOOKippZABatesEAda SilvaAATaylorLCMartinezGJet alLoss of hepatic PPARα in mice causes hypertension and cardiovascular disease. Am J Physiology-Regulatory, Integr Comp Physiol (2023) 325(1):R81–R95. 10.1152/ajpregu.00057.2023
139.
MartiniPHogeSBenenatoKPresnyakVMcFadyenIKumarasingheESet alPolynucleotides encoding citrin or the treatment of citrullinemia type 2 United States: Modernatx, Inc.Cambridge, MA (US) (2023). [updated 17-07-202312-07-2023]. 307].
140.
UnitaSHirashimaNShimadaMTsunekawaTTanakaDKondoTet alSuccessful treatment of adult-onset type II citrullinemia with a low-carbohydrate diet and L-arginine after DNA analysis produced a definitive diagnosis. Clin J Gastroenterol (2020) 13:823–33. 10.1007/s12328-019-01083-6
141.
KobayashiKSinasacDSIijimaMBorightAPBegumLLeeJRet alThe gene mutated in adult-onset type II citrullinaemia encodes a putative mitochondrial carrier protein. Nat Genet (1999) 22(2):159–63. 10.1038/9667
142.
SahekiTInoueKOnoHFujimotoYFuruieSYamamuraK-iet alOral aversion to dietary sugar, ethanol and glycerol correlates with alterations in specific hepatic metabolites in a mouse model of human citrin deficiency. Mol Genet Metab (2017) 120(4):306–16. 10.1016/j.ymgme.2017.02.004
143.
SpiritosZSalvadorSMosqueraDWilderJ. Acute intermittent porphyria: current perspectives and case presentation. Ther Clin Risk Management (2019) 15:1443–51. 10.2147/tcrm.s180161
144.
KizilaslanEZGhadgeNMMartinezABassMWinayakRMathewMet alAcute intermittent porphyria's symptoms and management: a narrative review. Cureus (2023) 15(3):e36058. 10.7759/cureus.36058
145.
RamanujamVSAndersonKE. Porphyria diagnostics-part 1: a brief overview of the porphyrias. Curr Protoc Hum Genet (2015) 86:17.20.1–17.20.6. 10.1002/0471142905.hg1720s86
146.
MehtaMRathGPPadhyUPMardaMDashHDashHH. Intensive care management of patients with acute intermittent porphyria: clinical report of four cases and review of literature. Indian J Crit Care Med (2010) 14(2):88–91. 10.4103/0972-5229.68222
147.
PischikEKauppinenR. An update of clinical management of acute intermittent porphyria. The Appl Clin Genet (2015) 8:201–14. 10.2147/tacg.s48605
148.
FloderusYSardhEMöllerCAnderssonCRejkjaerLAnderssonDEet alVariations in porphobilinogen and 5-aminolevulinic acid concentrations in plasma and urine from asymptomatic carriers of the acute intermittent porphyria gene with increased porphyrin precursor excretion. Clin Chem (2006) 52(4):701–7. 10.1373/clinchem.2005.058198
149.
GerischerLMScheibeFNümannAKöhnleinMStölzelUMeiselA. Acute porphyrias - a neurological perspective. Brain Behav (2021) 11(11):e2389. 10.1002/brb3.2389
150.
Valbuena ValecillosAYathamPAldermanMShapiroLTiozzoEGoberJ. Acute intermittent porphyria: a review and rehabilitation perspective. Cureus (2023) 15(8):e44260. 10.7759/cureus.44260
151.
AjayiTWardRSummersBByrnsJKappusMChoiSet alPathophysiology, pharmacology and treatment of acute intermittent porphyria: a patient case description and recommendations from the current literature. J Exploratory Res Pharmacol (2017) 2(2):49–53. 10.14218/jerp.2016.00022
152.
StölzelUDossMOSchuppanD. Clinical guide and update on porphyrias. Gastroenterology (2019) 157(2):365–81.e4. 10.1053/j.gastro.2019.04.050
153.
LongoMPaoliniEMeroniMDongiovanniP. Cutting-edge therapies and novel strategies for acute intermittent porphyria: step-By-step towards the solution. Biomedicines (2022) 10(3):648. 10.3390/biomedicines10030648
154.
MartiniPHogeSBenenatoKPresnvakVJiangIainL. Polynucleotides encoding porphobilinogen deaminase for the treatment of acute intermittent porphyria United States (2017). Available online at: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2017201346&_cid=P10-M8FOQP-49774-1 (Accessed November 23, 2017).
155.
BrusilowSWHorwichAL. Urea cycle enzymes. In: ValleDLAntonarakisSBallabioABeaudet al.MitchellGA, editors. The online metabolic and molecular bases of inherited disease. New York, NY: McGraw-Hill Education (2019).
156.
QuinonezSCLeeKN. Citrullinemia type I. In: AdamMPFeldmanJMirzaaGMPagonRAWallaceSEAmemiyaA, editors. GeneReviews(®). Seattle (WA): University of Washington, Seattle copyright © 1993-2025, university of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved (2004).
157.
JessGThoeneM. Rare disease database. Natl Organ Rare Disord (2021).
158.
KangHKimMLeeJH. Nutritional management in a patient with citrullinemia type 1. Clin Nutr Res (2021) 10(3):268–77. 10.7762/cnr.2021.10.3.268
159.
HeartleinMDeRosaFBoeglinL. mRNA therapy for argininosuccinate synthetase deficiency (2022).
160.
CollinsF. Cystic fibrosis: molecular biology and therapeutic implications. Science (1992) 256(5058):774–9. 10.1126/science.1375392
161.
KeremBRommensJMBuchananJAMarkiewiczDCoxTKChakravartiAet alIdentification of the cystic fibrosis gene: genetic analysis. Science (1989) 245(4922):1073–80. 10.1126/science.2570460
162.
BearCELiCHKartnerNBridgesRJJensenTJRamjeesinghMet alPurification and functional reconstitution of the cystic fibrosis transmembrane conductance regulator (CFTR). Cell (1992) 68(4):809–18. 10.1016/0092-8674(92)90155-6
163.
ChenQShenYZhengJ. A review of cystic fibrosis: basic and clinical aspects. Anim models Exp Med (2021) 4(3):220–32. 10.1002/ame2.12180
164.
ElbornJS. Cystic fibrosis. The Lancet (2016) 388(10059):2519–31. 10.1016/s0140-6736(16)00576-6
165.
OngTRamseyBW. Cystic fibrosis: a review. JAMA (2023) 329(21):1859–71. 10.1001/jama.2023.8120
166.
BenenatoKHogeSMcFadyenIPresnyakVMartiniPKumarasingheES. Polynucleotides encoding cystic fibrosis transmembrane conductance regulator for the treatment of cystic fibrosis (2019).
167.
FabryJ. Ein Beitrag zur Kenntniss der Purpura haemorrhagica nodularis (Purpura papulosa haemorrhagica Hebrae). Arch F Dermat (1898) 43:187–200. 10.1007/bf01986897
168.
WannerCAradMBaronRBurlinaAElliottPMFeldt-RasmussenUet alEuropean expert consensus statement on therapeutic goals in fabry disease. Mol Genet Metab (2018) 124(3):189–203. 10.1016/j.ymgme.2018.06.004
169.
Ilie-RobertDŞtefan-GeorgeF. Fabry disease - current data and therapeutic approaches. Rom J Morphol Embryol (2021) 62(1):5–11. 10.47162/rjme.62.1.01
170.
GermainDP. Fabry disease. Orphanet J rare Dis (2010) 5:30. 10.1186/1750-1172-5-30
171.
TuttolomondoASimonettaIDuroGPecoraroRMiceliSColombaPet alInter-familial and intra-familial phenotypic variability in three Sicilian families with anderson-fabry disease. Oncotarget (2017) 8(37):61415–24. 10.18632/oncotarget.18250
172.
MeiklePJHopwoodJJClagueAECareyWF. Prevalence of lysosomal storage disorders. JAMA (1999) 281(3):249–54. 10.1001/jama.281.3.249
173.
MurrayGJAnverMRKennedyMAQuirkJMSchiffmannR. Cellular and tissue distribution of intravenously administered agalsidase alfa. Mol Genet Metab (2007) 90(3):307–12. 10.1016/j.ymgme.2006.11.008
174.
BradyROSchiffmannR. Clinical features of and recent advances in therapy for fabry disease. JAMA (2000) 284(21):2771–5. 10.1001/jama.284.21.2771
175.
YuasaTTakenakaTHiguchiKUchiyamaNHorizoeYCyaenHet alFabry disease. J Echocardiography. (2017) 15(4):151–7. 10.1007/s12574-017-0340-x
176.
SchiffmannRWarnockDGBanikazemiMBultasJLinthorstGEPackmanSet alFabry disease: progression of nephropathy, and prevalence of cardiac and cerebrovascular events before enzyme replacement therapy. Nephrol Dial Transplant (2009) 24(7):2102–11. 10.1093/ndt/gfp031
177.
LeeKJinXZhangKCopertinoLAndrewsLBaker-MalcolmJet alA biochemical and pharmacological comparison of enzyme replacement therapies for the glycolipid storage disorder fabry disease. Glycobiology. (2003) 13(4):305–13. 10.1093/glycob/cwg034
178.
SakurabaHMurata-OhsawaMKawashimaITajimaYKotaniMOhshimaTet alComparison of the effects of agalsidase alfa and agalsidase beta on cultured human fabry fibroblasts and Fabry mice. J Hum Genet (2006) 51(3):180–8. 10.1007/s10038-005-0342-9
179.
FelisAWhitlowMKrausAWarnockDGWallaceE. Current and investigational therapeutics for fabry disease. Kidney Int Rep (2020) 5(4):407–13. 10.1016/j.ekir.2019.11.013
180.
ZhuXYinLTheisenMZhuoJSiddiquiSLevyBet alSystemic mRNA therapy for the treatment of fabry disease: preclinical studies in wild-type mice, fabry mouse model, and wild-type non-human Primates. The Am J Hum Genet (2019) 104(4):625–37. 10.1016/j.ajhg.2019.02.003
181.
Al HafidNChristodoulouJ. Phenylketonuria: a review of current and future treatments. Translational Pediatr (2015) 4(4):304–17. 10.3978/j.issn.2224-4336.2015.10.07
182.
van SpronsenFJBlauNHardingCBurlinaALongoNBoschAM. Phenylketonuria. Nat Rev Dis primers (2021) 7(1):36. 10.1038/s41572-021-00267-0
183.
KaufmanS. The phenylalanine hydroxylating system from mammalian liver. Adv Enzymol Relat Areas Mol Biol (1971) 35:245–319. 10.1002/9780470122808.ch6
184.
GarbadeSFShenNHimmelreichNHaasDTrefzFKHoffmannGFet alAllelic phenotype values: a model for genotype-based phenotype prediction in phenylketonuria. Genet Med (2019) 21(3):580–90. 10.1038/s41436-018-0081-x
185.
AcostaPBYannicelliSSinghREisasLJ2ndKennedyMJBernsteinLet alIntake and blood levels of fatty acids in treated patients with phenylketonuria. J Pediatr Gastroenterol Nutr (2001) 33(3):253–9. 10.1002/j.1536-4801.2001.tb07453.x
186.
AcostaPBYannicelliSSinghRMofidiSSteinerRDeVincentisEet alNutrient intakes and physical growth of children with phenylketonuria undergoing nutrition therapy. J Am Diet Assoc (2003) 103(9):1167–73. 10.1016/s0002-8223(03)00983-0
187.
AcostaPBYannicelliSSinghRHElsasLJ2ndMofidiSSteinerRD. Iron status of children with phenylketonuria undergoing nutrition therapy assessed by transferrin receptors. Genet Med (2004) 6(2):96–101. 10.1097/01.gim.0000117335.50541.f3
188.
FrankDRMHDiasA. mRNA therapy for phenylketonuria (2022).
189.
KohlerLPRPuertollanoRRabenN. Pompe disease: from basic science to therapy. Neurotherapeutics (2018) 15(4):928–42. 10.1007/s13311-018-0655-y
190.
TavernaSCGCammarataGColombaPSciarrinoSZizzoCFrancofonteDet alPompe disease: pathogenesis, molecular genetics and diagnosis. Aging (2020) 12(15):15856–74. 10.18632/aging.103794
191.
Borie-GuichotMTMTranMLGénissonYBallereauSDehouxC. Pharmacological chaperone therapy for pompe disease. Molecules (2021) 26(23):7223. 10.3390/molecules26237223
192.
StevensDM-NSMilani-NejadSMozaffarT. Pompe disease: a clinical, diagnostic, and therapeutic overview. Curr Treat Options Neurol (2022) 24(11):573–88. 10.1007/s11940-022-00736-1
193.
SchoserB. Pompe disease: what are we missing?Ann Transl Med (2019) 7(13):292. 10.21037/atm.2019.05.29
194.
ReuserAJJvdPAPloegATChienYLlerenaJJrAbbottMClemensPRet alGAA variants and phenotypes among 1,079 patients with pompe disease: data from the pompe registry. Hum Mutat (2019) 40(11):2146–64. 10.1002/humu.23934
195.
BellottiASALAndreoliLRonchiDBresolinNComiGPCortiS. Molecular approaches for the treatment of pompe disease. Mol Neurobiol (2020) 57(2):1259–80. 10.1007/s12035-019-01820-5
196.
AusemsMGVJVerbiestJHermansMKroosMBeemerFWokkeJet alFrequency of glycogen storage disease type II in the Netherlands: implications for diagnosis and genetic counselling. Eur J Hum Genet (1999) 7(6):713–6. 10.1038/sj.ejhg.5200367
197.
MartiniukFCAChenAMackAArvanitopoulosEChenYRomWNet alCarrier frequency for glycogen storage disease type II in New York and estimates of affected individuals born with the disease. Am J Med Genet (1998) 79(1):69–72. 10.1002/(sici)1096-8628(19980827)79:1<69::aid-ajmg16>3.0.co;2-k
198.
KornfeldS. Trafficking of lysosomal enzymes in normal and disease states. J Clin Invest (1986) 77(1):1–6. 10.1172/jci112262
199.
WisselaarHAKMKroosMAHermansMMvan BeeumenJReuserAJ. Structural and functional changes of lysosomal acid alpha-glucosidase during intracellular transport and maturation. J Biol Chem (1993) 268(3):2223–31.
200.
LimJALLLiLRabenN. Pompe disease: from pathophysiology to therapy and back again. Front Aging Neurosci (2014) 6:177. 10.3389/fnagi.2014.00177
201.
van der PloegATReuserAJ. Pompe's disease. The Lancet (2008) 372(9646):1342–53. 10.1016/s0140-6736(08)61555-x
202.
KishnaniPSCDCorzoDLeslieNDGruskinDVan der PloegAClancyJPet alEarly treatment with alglucosidase alpha prolongs long-term survival of infants with pompe disease. Pediatr Res (2009) 66(3):329–35. 10.1203/PDR.0b013e3181b24e94
203.
van GelderCMH-WMHoogeveen-WesterveldMKroosMAPlugIvan der PloegATReuserAJJ. Enzyme therapy and immune response in relation to CRIM status: the Dutch experience in classic infantile pompe disease. J Inherit Metab Dis (2015) 38(2):305–14. 10.1007/s10545-014-9707-6
204.
Frank DeRosaMH. mRNA therapy for pompe disease (2023).
205.
A Phase 3b, Open-label, safety and immunogenicity study of SARS-CoV-2 mRNA-1273 vaccine in adult solid organ transplant recipients and healthy controls (2021). Available online at: https://clinicaltrials.gov/study/NCT04860297 (Accessed April 16 2024).
206.
A phase 3, randomized, observer-blind study to evaluate the safety, tolerability, and immunogenicity of multiple formulations of the vaccine candidate bnt162b2 against covid 19 in healthy adults 18 through 55 years of age 2021 (2021). Available online at: https://clinicaltrials.gov/study/NCT04816669 (Accessed November 30, 2022).
207.
Safety and immunogenicity of SARS-CoV-2 mRNA vaccine (BNT162b1) in Chinese healthy subjects: a phase I, randomized, placebo-controlled, observer-blind study 2020 (2020). Available online at: https://clinicaltrials.gov/study/NCT04523571 (Accessed July 28, 2024).
208.
LiJHuiAZhangXYangYTangRYeHet alSafety and immunogenicity of the SARS-CoV-2 BNT162b1 mRNA vaccine in younger and older Chinese adults: a randomized, placebo-controlled, double-blind phase 1 study. Nat Med (2021) 27(6):1062–70. 10.1038/s41591-021-01330-9
209.
A Global, Multi-center, randomized, double-blind, placebo-controlled, phase III clinical study to evaluate the protective efficacy, safety and immunogenicity of SARS-CoV-2 messenger ribonucleic acid (mRNA) vaccine in population aged 18 years and older (2021). Available online at: https://clinicaltrials.gov/study/NCT04847102 (Accessed November 10, 2024).
210.
A Phase 2 Randomized, Observer-blind, placebo-controlled study to assess the safety, reactogenicity, and immunogenicity of the SARS CoV-2 vaccine ARCT-021 in healthy adult participants (2020). Available online at: https://clinicaltrials.gov/study/NCT04668339 (Accessed January 07, 2021).
211.
Immunogenicity and safety of the first-in-human SARS-CoV-2 mRNA vaccine formulation in healthy adults 18 years of age and older (2021). Available online at: https://clinicaltrials.gov/study/NCT04798027 (Accessed December 13, 2024).
212.
A phase 1/2, dose-finding study to evaluate safety, tolerability, and immunogenicity of the ChulaCov19 vaccine in healthy adults (2020). Available online at: https://clinicaltrials.gov/study/NCT04566276 (Accessed May 03, 2022).
213.
PuthanakitTPrompetcharaEGatechompolSKetloyCThitithanyanontAJongkaewwattanaAet alPhase II prefusion non-stabilised Covid-19 mRNA vaccine randomised study. Scientific Rep (2024) 14(1):2373. 10.1038/s41598-023-49653-6
214.
Phase I safety and immunogenicity trial of an investigational RNActive® rabies vaccine (CV7201) in healthy adults (2014). Available online at: https://clinicaltrials.gov/study/NCT02241135 (Accessed October 25, 2022).
215.
AlbererMGnad-VogtUHongHSMehrKTBackertLFinakGet alSafety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. The Lancet (2017) 390(10101):1511–20. 10.1016/s0140-6736(17)31665-3
216.
A Non-Randomized, Open label, controlled, dose-escalation, phase I clinical trial to evaluate the safety, reactogenicity and immunogenicity of one or two administrations of candidate rabies mRNA vaccine CV7202 in healthy adult subjects (2018). Available online at: https://clinicaltrials.gov/study/NCT03713086 (Accessed October 12, 2024).
217.
AldrichCLeroux–RoelsIHuangKBBicaMALoeligerESchoenborn-KellenbergerOet alProof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: a phase 1 trial. Vaccine (2021) 39(8):1310–8. 10.1016/j.vaccine.2020.12.070
218.
LeeITNachbagauerREnszDSchwartzHCarmonaLSchaefersKet alSafety and immunogenicity of a phase 1/2 randomized clinical trial of a quadrivalent, mRNA-based seasonal influenza vaccine (mRNA-1010) in healthy adults: interim analysis. Nat Commun (2023) 14(1):3631. 10.1038/s41467-023-39376-7
219.
AnanworanichJLeeITEnszDCarmonaLSchaefersKAvanesovAet alSafety and immunogenicity of mRNA-1010, an investigational seasonal influenza vaccine. In: Healthy adults: final results from a phase 1/2 randomized trial (2024). 10.1093/infdis/jiae329
220.
Phase 1/2, randomized, observer-blind, dose-ranging study to evaluate the safety, reactogenicity, and immunogenicity of mRNA-1020 and mRNA-1030 candidate seasonal influenza vaccines in healthy adults (2022). Available online at: https://clinicaltrials.gov/study/NCT05333289 (Accessed April 06 2022).
221.
A phase 2a randomized, observer-blind, dose-finding study to evaluate the immunogenicity and safety of mRNA-based multivalent seasonal influenza vaccine candidates in adults 18 years of age and older (2024). Available online at: https://clinicaltrials.gov/study/NCT06431607 (Accessed May 23, 2024).
222.
A phase 1/2, randomized, dose-Finding/dose-confirmation study to evaluate the reactogenicity, safety and immunogenicity of mRNA-based multivalent seasonal influenza vaccine candidates administered in healthy younger and older adults (2023). Available online at: https://clinicaltrials.gov/study/NCT05823974 (Accessed February 27, 2023).
223.
A phase 1, randomized, placebo-controlled, dose ranging study to evaluate the safety, tolerability, pharmacokinetics, and pharmacodynamics of mRNA-1944, encoding for an anti-chikungunya virus monoclonal antibody, in healthy adults (2019). Available online at: https://clinicaltrials.gov/study/NCT03829384 (Accessed January 22, 2023).
224.
A phase 1, randomized, open-label clinical trial to evaluate the safety and immunogenicity of BG505 MD39.3, BG505 MD39.3 gp151, and BG505 MD39.3 gp151 CD4KO HIV trimer mRNA vaccines in healthy, HIV-uninfected adult participants (2021). Available online at: https://clinicaltrials.gov/study/NCT05217641 (Accessed February 11, 2022).
225.
A phase 1, randomized, first-in-human, open-label study to evaluate the safety and immunogenicity of eOD-GT8 60mer mRNA vaccine (mRNA-1644) and Core-g28v2 60mer mRNA vaccine (mRNA-1644v2-Core) in HIV-1 uninfected adults in good general health. International AIDS Vaccine Initiative Active (2021). Available online at: https://clinicaltrials.gov/study/NCT05001373 (Accessed April 29, 2024).
226.
A phase 1 clinical trial to evaluate the safety and immunogenicity of ferritin nanoparticles expressing native-like HIV-1 envelope trimers followed by boost with mRNA lipid nanoparticles encoding a native-like HIV-1 envelope trimer in adults without HIV. National Institute of Allergy and Infectious Diseases NIAID (2023). Available online at: https://clinicaltrials.gov/study/NCT05903339 (Accessed August 29, 2023).
227.
LorentzenCLHaanenJBMetÖSvaneIM. Clinical advances and ongoing trials of mRNA vaccines for cancer treatment (2022) 23(10):e450–e8.
228.
FierroCBruneDShawMSchwartzHKnightlyCLinJet alSafety and immunogenicity of a messenger RNA–based cytomegalovirus vaccine in healthy adults: results from a phase 1 randomized clinical trial. The J Infect Dis (2024) 230(3):e668–e78.
229.
A phase 1, randomized, observer-blind, placebo-controlled, dose-ranging study to evaluate the safety, reactogenicity, and immunogenicity of cytomegalovirus vaccines mRNA-1647 and mRNA-1443 when administered to healthy adults (2017). Available online at: https://clinicaltrials.gov/study/NCT03382405 (Accessed November 16, 2023).
230.
A phase 1/2, randomized, observer-blind, active-controlled, dose-ranging study to evaluate the safety, reactogenicity and immunogenicity of mRNA-1468, a candidate vaccine to prevent herpes zoster (HZ) in healthy adults ≥50 years of age (2023). Available online at: https://clinicaltrials.gov/study/NCT05701800 (Accessed January 23, 2023).
231.
A phase 1/2 randomized, observer-blind study to evaluate the safety, tolerability, and immunogenicity of a modified rna vaccine against Varicella zoster virus in healthy individual (2023). Available online at: https://clinicaltrials.gov/study/NCT05703607 (Accessed January 25, 2023).
232.
AugustAShawCALeeHKnightlyCKalidindiaSChuL, editors. Safety and immunogenicity of an mRNA-based human metapneumovirus and parainfluenza virus type 3 combined vaccine in healthy adults. Open forum infectious diseases. Oxford University Press (2022).
233.
A phase 1, randomized, observer-blind, placebo-controlled, dose-ranging study to evaluate the safety, reactogenicity, and immunogenicity of mRNA-1653, a combined human metapneumovirus and human parainfluenza virus type 3 vaccine, when administered to healthy adults (2018). Available online at: https://clinicaltrials.gov/study/NCT03392389 (Accessed December 04, 2023).
234.
JansenYKruseVCorthalsJSchatsKvan DamPJSeremetTet alA randomized controlled phase II clinical trial on mRNA electroporated autologous monocyte-derived dendritic cells (TriMixDC-MEL) as adjuvant treatment for stage III/IV melanoma patients who are disease-free following the resection of macrometastases. Cancer Immunol Immunother (2020) 69(12):2589–98. 10.1007/s00262-020-02618-4
235.
Randomized controlled phase II clinical trial on mRNA electroporated autologous dendritic cells for stage III/IV melanoma patients who are disease-free following the local treatment of macrometastases (2012). Available online at: https://clinicaltrials.gov/study/NCT01676779 (Accessed November 25, 2023).
236.
AlsalloumAShevchenkoJASennikovS. NY‐ESO‐1 antigen: a promising frontier in cancer immunotherapy. Clin Translational Med (2024) 14(9):e70020. 10.1002/ctm2.70020
237.
Open-label, randomized phase II trial with BNT111 and cemiplimab in combination or as single agents in patients with Anti-PD-1-refractory/Relapsed, unresectable stage III or IV melanoma (2020). Available online at: https://clinicaltrials.gov/study/NCT04526899 (Accessed May 19, 2024).
238.
SchmidtMVoglerIDerhovanessianEOmokokoTGodehardtEAttigSet al88MO T-cell responses induced by an individualized neoantigen specific immune therapy in post (neo) adjuvant patients with triple negative breast cancer. Ann Oncol (2020) 31:S276. 10.1016/j.annonc.2020.08.209
239.
PapachristofilouAHippMMKlinkhardtUFrühMSebastianMWeissCet alPhase Ib evaluation of a self-adjuvanted protamine formulated mRNA-based active cancer immunotherapy, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non-small cell lung cancer. J ImmunoTherapy Cancer (2019) 7(1):38. 10.1186/s40425-019-0520-5
240.
A phase 1/2 study of combination immunotherapy and mRNA vaccine in subjects with non-small cell lung cancer (NSCLC). Ludwig Institute for Cancer Research (2017). Available online at: https://clinicaltrials.gov/study/NCT03164772.
241.
ShimKJoHJeoungD. Cancer/testis antigens as targets for RNA-based anticancer therapy. Int J Mol Sci (2023) 24(19):14679. 10.3390/ijms241914679
242.
A phase 1, open-label, multicenter study to assess the safety and tolerability of mRNA-5671/V941 as a monotherapy and in combination with pembrolizumab in participants with KRAS mutant advanced or metastatic non-small cell lung cancer, colorectal cancer or pancreatic adenocarcinoma (2019). Available online at: https://clinicaltrials.gov/study/NCT03948763 (Accessed June 26, 2023).
243.
A multi-site, open-label, phase II, randomized, controlled trial to compare the efficacy of RO7198457 versus watchful waiting in resected, stage II (high risk) and stage III colorectal cancer patients who are ctDNA positive following resection (2020). Available online at: https://clinicaltrials.gov/study/NCT04486378 (Accessed May 08, 2024).
244.
BhattKHNellerMASrihariSCrooksPLekieffreLAftabBTet alProfiling HPV-16–specific T cell responses reveals broad antigen reactivities in oropharyngeal cancer patients. J Exp Med (2020) 217(10):e20200389. 10.1084/jem.20200389
245.
An open label phase II randomized trial of BNT113 in combination with pembrolizumab versus pembrolizumab monotherapy as a first line therapy in patients with unresectable recurrent, or metastatic head and neck squamous cell carcinoma (HNSCC) which is positive for human papilloma virus 16 (HPV16+) and expresses PD-L1 (2020). Available online at: https://clinicaltrials.gov/study/NCT04534205 (Accessed January 07, 2024).
246.
RosenbergAJPerezCAGuoWde Oliveira NovaesJMda Silva ReisKFOMcGarrahPWet alBreaking ground in recurrent or metastatic head and neck squamous cell carcinoma: novel therapies beyond PD-L1 immunotherapy. Am Soc Clin Oncol Educ Book (2024) 44(3):e433330. 10.1200/edbk_433330
247.
MackensenAHaanenJKoeneckeCAlsdorfWWagner-DrouetEHeudoblerDet alLBA38 BNT211-01: a phase I trial to evaluate safety and efficacy of CLDN6 CAR T cells and CLDN6-encoding mRNA vaccine-mediated in vivo expansion in patients with CLDN6-positive advanced solid tumours. Ann Oncol (2022) 33:S1404–S1405. 10.1016/j.annonc.2022.08.035
248.
Clinicaltrials. Phase I/IIa, first-in-human (FIH), open-label, dose escalation trial with expansion cohorts to evaluate safety and preliminary efficacy of CLDN6 CAR-T with or without CLDN6 RNA-LPX in patients with CLDN6-positive relapsed or refractory advanced solid tumors. BioNTech Cell & Gene Therapies GmbH (2020). Available online at: https://clinicaltrials.gov/study/NCT04503278.
249.
GainorJFPatelMRWeberJGutierrezMBaumanJEClarkeJMet al1530 T-cell responses to individualized neoantigen therapy (INT) mRNA-4157 (V940) as monotherapy or in combination with pembrolizumab. Late-Breaking Abstr (2023) A1754. 10.1136/jitc-2023-sitc2023.1530
250.
BurrisHAPatelMRChoDCClarkeJMGutierrezMZaksTZet alA phase I multicenter study to assess the safety, tolerability, and immunogenicity of mRNA-4157 alone in patients with resected solid tumors and in combination with pembrolizumab in patients with unresectable solid tumors. J Clin Oncol (2019) 37(15_Suppl. l):2523. 10.1200/jco.2019.37.15_suppl.2523
251.
Clinicaltrials. A phase 1, open-label, multicenter study to assess the safety, tolerability, and immunogenicity of mRNA-4157 alone and in combination in participants with solid tumors. ModernaTX, Inc. (2017). Available online at: https://clinicaltrials.gov/study/NCT03313778.
252.
Clinicaltrials. A phase 3, randomized, double-blind, Placebo- and active-comparator-controlled clinical study of adjuvant V940 (mRNA-4157) plus pembrolizumab versus adjuvant placebo plus pembrolizumab in participants with high-risk stage II-IV melanoma (INTerpath-001). Merck Sharp & Dohme LLC (2023). Available online at: https://clinicaltrials.gov/study/NCT05933577.
253.
Clinicaltrials. A phase 3, randomized, double-blind, Placebo- and active-comparator-controlled clinical study of adjuvant V940 (mRNA-4157) plus pembrolizumab versus adjuvant placebo plus pembrolizumab in participants with resected stage II, IIIA, IIIB (N2) non-small cell lung cancer (INTerpath-002). Merck Sharp & Dohme LLC (2023). Available online at: https://clinicaltrials.gov/study/NCT06077760.
254.
Clinicaltrials. Phase 1 clinical trial of personalized neoantigen tumor vaccines and programmed death-ligand 1 (PD-L1) blockade in patients with surgically resected pancreatic cancer. Memorial Sloan Kettering Cancer Center (2019). Available online at: https://clinicaltrials.gov/study/NCT04161755.
255.
RojasLASethnaZSoaresKCOlceseCPangNPattersonEet alPersonalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature (2023) 618(7963):144–50. 10.1038/s41586-023-06063-y
256.
CafriGGartnerJJHopsonKMeehanRSZaksTZRobbinsPet alImmunogenicity and tolerability of personalized mRNA vaccine mRNA-4650 encoding defined neoantigens expressed by the autologous cancer. American Society of Clinical Oncology (2019).
257.
CafriGGartnerJJZaksTHopsonKLevinNPariaBCet almRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J Clin Invest (2020) 130(11):5976–88. 10.1172/jci134915
258.
Clinicaltrials. A global, phase 1/2, open label, dose escalation study to evaluate the safety, pharmacodynamics, and pharmacokinetics of mRNA-3704 in patients with isolated methylmalonic acidemia due to Methylmalonyl-CoA mutase deficiency. ModernaTX, Inc. (2019). Available online at: https://clinicaltrials.gov/study/NCT03810690.
259.
Clinicaltrials. A global, phase 1/2, open-label, dose optimization study to evaluate the safety, pharmacodynamics, and pharmacokinetics of mRNA-3927 in participants with propionic acidemia. ModernaTX, Inc. (2019). Available online at: https://clinicaltrials.gov/study/NCT04159103.
260.
Clinicaltrials. Phase 2, randomized, double-blind, placebo-controlled, nested single and multiple ascending dose study to evaluate the safety, tolerability and pharmacokinetics of ARCT-810 in adolescent and adult participants with ornithine transcarbamylase deficiency. Arcturus Therapeutics, Inc. (2022). Available online at: https://clinicaltrials.gov/study/NCT05526066.
261.
Seker YilmazBGissenP. Genetic therapy approaches for ornithine transcarbamylase deficiency. Biomedicines (2023) 11(8):2227. 10.3390/biomedicines11082227
262.
BaiXChenQLiFTengYTangMHuangJet alOptimized inhaled LNP formulation for enhanced treatment of idiopathic pulmonary fibrosis via mRNA-mediated antibody therapy. Nat Commun (2024) 15(1):6844. 10.1038/s41467-024-51056-8
263.
Clinicaltrials. A phase 1/2 dose escalation study evaluating the safety, and tolerability and efficacy of VX-522 in subjects 18 years of age and older with cystic fibrosis and a CFTR genotype not responsive to CFTR modulator therapy. Vertex Pharmaceuticals Incorporated (2022). Available online at: https://clinicaltrials.gov/study/NCT05668741.
264.
RoweSMZuckermanJBDorganDLascanoJMcCoyKJainMet alInhaled mRNA therapy for treatment of cystic fibrosis: interim results of a randomized, double-blind, placebo-controlled phase 1/2 clinical study. J Cystic Fibrosis (2023) 22(4):656–64. 10.1016/j.jcf.2023.04.008
265.
Clinicaltrials. A phase 1/2, randomized, double-blinded, placebo-controlled, combined single and multiple ascending dose study evaluating the safety, tolerability, and biological activity of MRT5005 administered by nebulization to adult subjects with cystic fibrosis. Translate Bio, Inc. (2017). Available online at: https://clinicaltrials.gov/study/NCT03375047.
266.
GanL-MLagerström-FermérMCarlssonLGArfvidssonCEgnellA-CRudvikAet alIntradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat Commun (2019) 10(1):871. 10.1038/s41467-019-08852-4
267.
Clinicaltrials. A phase I, randomized, single-blind, placebo controlled study to assess the safety, tolerability and pharmacodynamics of AZD8601 after single dose administration to Male patients with type II diabetes mellitus (T2DM). AstraZeneca (2016). Available online at: https://clinicaltrials.gov/study/NCT02935712.
268.
CollénABergenhemNCarlssonLChienKRHogeSGanL-Met alVEGFA mRNA for regenerative treatment of heart failure. Nat Rev Drug Discov (2022) 21(1):79–80. 10.1038/s41573-021-00355-6
269.
AnttilaVSarasteAKnuutiJHedmanMJaakkolaPLaugwitzK-Let alDirect intramyocardial injection of VEGF mRNA in patients undergoing coronary artery bypass grafting. Mol Ther (2023) 31(3):866–74. 10.1016/j.ymthe.2022.11.017
270.
Clinicaltrials. A randomized, double-blind, placebo-controlled, multi-centre, sequential design, phase IIa study to evaluate safety and tolerability of epicardial injections of AZD8601 during coronary artery bypass grafting surgery. AstraZeneca (2017). Available online at: https://clinicaltrials.gov/study/NCT03370887.
271.
GillmoreJDGaneETaubelJKaoJFontanaMMaitlandMLet alCRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. New Engl J Med (2021) 385(6):493–502. 10.1056/nejmoa2107454
272.
Clinicaltrials. Phase 1 two-part (Open-label, single ascending dose (part 1) and open-label, single dose expansion (part 2)) study to evaluate safety, tolerability, pharmacokinetics, and pharmacodynamics of NTLA-2001 in patients with hereditary transthyretin amyloidosis with polyneuropathy (ATTRv-PN) and patients with transthyretin amyloidosis-related cardiomyopathy (ATTR-CM). Intellia Therapeutics (2020). Available online at: https://clinicaltrials.gov/study/NCT04601051.
273.
Clinicaltrials. MAGNITUDE: a phase 3, multinational, multicenter, randomized, double-blind, placebo-controlled study to evaluate the efficacy and safety of NTLA-2001 in participants with transthyretin amyloidosis with cardiomyopathy (ATTR-CM). Intellia Therapeutics (2023). Available online at: https://clinicaltrials.gov/study/NCT06128629.
274.
Analysis of off-target effects and risk assessment leading from preclinical to clinical trials of gene-edited therapeutic products. Ther Innovation & Regul Sci (2023) 57(3):538–51. 10.1007/s43441-022-00481-2
275.
GaoMZhangQFengXHLiuJ. Synthetic modified messenger RNA for therapeutic applications. Acta Biomater (2021) 131:1–15. 10.1016/j.actbio.2021.06.020
276.
SunNNingBHanssonKMBruceACSeamanSAZhangCet alModified VEGF-A mRNA induces sustained multifaceted microvascular response and accelerates diabetic wound healing. Scientific Rep (2018) 8(1):17509. 10.1038/s41598-018-35570-6
277.
ZhaWWangJGuoZZhangYWangYDongSet alEfficient delivery of VEGF-A mRNA for promoting diabetic wound healing via ionizable lipid nanoparticles. Int J Pharmaceutics (2023) 632:122565. 10.1016/j.ijpharm.2022.122565
278.
AnttilaVSarasteAKnuutiJJaakkolaPHedmanMSvedlundSet alSynthetic mRNA encoding VEGF-A in patients undergoing coronary artery bypass grafting: design of a phase 2a clinical trial. Mol Ther - Methods & Clin Development (2020) 18:464–72. 10.1016/j.omtm.2020.05.030
279.
ZhouTGeorgievIWuXYangZYDaiKFinziAet alStructural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science (2010) 329(5993):811–7. 10.1126/science.1192819
280.
PardiNSecretoAJShanXDeboneraFGloverJYiYet alAdministration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat Commun (2017) 8(1):14630. 10.1038/ncomms14630
281.
SchwartzMShnayderMNachshonAAraziTKitsbergYLevi SamiaRet alMolecular characterization of human cytomegalovirus infection with single-cell transcriptomics. Nat Microbiol (2023) 8(3):455–68. 10.1038/s41564-023-01325-x
282.
BalegamireSJMcClymontECroteauADodinPGanttSBesharatiAAet alPrevalence, incidence, and risk factors associated with cytomegalovirus infection in healthcare and childcare worker: a systematic review and meta-analysis. Syst Rev (2022) 11(1):131–14. 10.1186/s13643-022-02004-4
283.
GönczölEde TaisneCHirkaGBerencsiKLinWPaolettiEet alHigh expression of human cytomegalovirus (HCMV)-gB protein in cells infected with a vaccinia-gB recombinant: the importance of the gB protein in HCMV immunity. Vaccine (1991) 9(9):631–7. 10.1016/0264-410x(91)90187-b
284.
McVoyMALeeRSaccoccioFMHartikkaJSmithLRMahajanRet alA cytomegalovirus DNA vaccine induces antibodies that block viral entry into fibroblasts and epithelial cells. Vaccine (2015) 33(51):7328–36. 10.1016/j.vaccine.2015.10.078
285.
ChenS-JWangS-CChenY-C. Challenges, recent advances and perspectives in the treatment of human cytomegalovirus infections. Trop Med Infect Dis (2022) 7(12):439. 10.3390/tropicalmed7120439
286.
CiaramellaGJohnSMousaviK. Inventors; ModernaTX, inc. Cambridge (US), assignee. Human cytomegalovirus RNA vaccines. United States patent US (2022) 11. 484,590 B2.
287.
FowlerKMuchaJNeumannMLewandowskiWKaczanowskaMGrysMet alA systematic literature review of the global seroprevalence of cytomegalovirus: possible implications for treatment, screening, and vaccine development. BMC Public Health (2022) 22(1):1659–15. 10.1186/s12889-022-13971-7
288.
KhalilAHeathPJonesCSoeAVilleY, Royal College of Obstetricians and Gy, naecologistsCongenital cytomegalovirus infection: update on screening, diagnosis and treatment: scientific impact paper no. 56. BJOG: Int J Obstet Gynaecol (2025) 132:e42–e52. 10.1111/1471-0528.17966
289.
KschonsakMJohnsonMCSchellingRGreenEMRougéLHoHet alStructural basis for HCMV pentamer receptor recognition and antibody neutralization. Sci Adv (2022) 8(10):eabm2536. 10.1126/sciadv.abm2536
290.
CiaramellaGHuangEY-CBahlKZaksTHimansuSElbashirSM. Infectious disease vaccines United States: ModernaTX, Inc.Cambridge (2022).
291.
AkingbolaAAdegbesanAAdewoleOAdegokeKBensonAEJomboPAet alThe mRNA-1647 vaccine: a promising step toward the prevention of cytomegalovirus infection (CMV). Hum Vaccin & Immunother (2025) 21(1):2450045. 10.1080/21645515.2025.2450045
292.
ModernaTXI. A study to evaluate the efficacy, safety, and immunogenicity of mRNA-1647 cytomegalovirus (CMV) vaccine in allogenic hematopoietic cell transplantation (HCT) participants. NIH.Gov: ClinicalTrials.gov ID NCT05683457 (2025).
293.
RahimiGRahimiBPanahiMAbkhizSSaraygord-AfshariNMilaniMet alAn overview of Betacoronaviruses-associated severe respiratory syndromes, focusing on sex-type-specific immune responses. Int Immunopharmacology (2021) 92:107365. 10.1016/j.intimp.2021.107365
294.
CiaramellaGHimansuS. BetaCoronavirus mRNA vaccine United States: modernatx. Inc.Cambridge. (2021).
295.
ZhaoYHuangJZhangLChenSGaoJJiaoH. The global transmission of new coronavirus variants. Environ Res (2022) 206:112240. 10.1016/j.envres.2021.112240
296.
PrompetcharaEKetloyCPalagaT. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol (2020) 38(1):1–9. 10.12932/AP-200220-0772
297.
AhmadSWaheedYIsmailSBhattiSAbbasiSWMuhammadK. Structure-based virtual screening identifies multiple stable binding sites at the RecA domains of SARS-CoV-2 helicase enzyme. Molecules (2021) 26(5):1446. 10.3390/molecules26051446
298.
SharmaGVolgmanASMichosED. Sex differences in mortality from COVID-19 pandemic: are men vulnerable and women protected?JACC: Case Rep (2020) 2(9):1407–10. 10.1016/j.jaccas.2020.04.027
299.
KleinSLFlanaganKL. Sex differences in immune responses. Nat Rev Immunol (2016) 16(10):626–38. 10.1038/nri.2016.90
300.
MunsterVJKoopmansMVan DoremalenNVan RielDde WitE. A novel coronavirus emerging in China—Key questions for impact assessment. New Engl J Med (2020) 382(8):692–4. 10.1056/nejmp2000929
301.
MsemburiWKarlinskyAKnutsonVAleshin-GuendelSChatterjiSWakefieldJ. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature (2023) 613(7942):130–7. 10.1038/s41586-022-05522-2
302.
GarryRF. Lassa fever—the road ahead. Nat Rev Microbiol (2023) 21(2):87–96. 10.1038/s41579-022-00789-8
303.
GüntherSLenzO. Lassa virus. Crit Rev Clin Lab Sci (2004) 41(4):339–90. 10.1080/10408360490497456
304.
van LeeuwenLPde JongWDoornekampLvan GorpECWismansPJGoeijenbierM. Exotic viral hepatitis; a review on epidemiology, pathogenesis, and treatment. J Hepatol (2022) 77:1431–43. 10.1016/j.jhep.2022.06.031
305.
HastieKMSaphireEO. Lassa virus glycoprotein: stopping a moving target. Curr Opin Virol (2018) 31:52–8. 10.1016/j.coviro.2018.05.002
306.
LarsonRADaiDHosackVTTanYBolkenTCHrubyDEet alIdentification of a broad-spectrum arenavirus entry inhibitor. J Virol (2008) 82(21):10768–75. 10.1128/jvi.00941-08
307.
JasnyEPetschB. Lassa virus vaccine Germany. CureVac AG Tübingen (DE) (2023).
308.
ValianteNZaksTHuangEY-CCiaramellaG. Cancer vaccines United States: ModernaTX, inc. Cambridge (2016).
309.
Rami-PortaRBolejackVGirouxDJChanskyKCrowleyJAsamuraHet alThe IASLC lung cancer staging project: the new database to inform the eighth edition of the TNM classification of lung cancer. J Thorac Oncol (2014) 9(11):1618–24. 10.1097/jto.0000000000000334
310.
ArzmiMHPp Abdul MajeedAMuazu MusaRMohd RazmanMAGanH-SMohd KhairuddinIet alEpidemiology, detection and management of cancer: an overview. In: Deep learning in cancer diagnostics: a feature-based transfer learning evaluation (2023). p. 1–7.
311.
KallenK-JFotin-MleczekMGnad-VogtU. Composition and vaccine for treating lung cancer Germany: Curevac AG tübingen (DE) (2021). p. 239.
312.
AshburnTHopsonKKeatingKSennJSwensonCBretonBet alRNA cancer vaccines United States: modernatx. Inc. Cambridge.
313.
ValianteNAshburnTFlopsonK. RNA cancer vaccines United States: ModernaTX, inc. Cambridge (2019).
314.
ZhongSBretonBMcfadyenIHopsonKLuczkowVGarnaasM. Personalized cancer vaccine epitope selection United States of America: modernatx, Inc. Cambridge (2020).
315.
ChenGDanglMGehoDNicholsGZhongH. mRNA based gene expression for personalizing patient cancer therapy with an mdm2 antagonist Switzerland. F Hoffmann-la Roche Ag (2015).
316.
KallenK-JFotin-MleczekMGnad-VogtULanderT. Composition and vaccine for treating prostate cancer Germany: Curevac AG Tübingen DE (2016).
317.
HuangEY-CTseS-WIacovelliJMckinneyKHopsonK. Messenger ribonucleic acids for enhancing immune responses and methods of use thereof United States: ModernaTX, Inc.Cambridge (2020).
318.
FrederickJGurumurthySMishraABaiA. Methods and compositions for treating cancer using mRNA therapeutics United States: ModernaTX, Inc.Cambridge (2023).
319.
FrederickJPHewittSBaiAHogeSGMcfadyenVPBenenatoKet almRNA combination therapy for the treatment of cancer United States: ModernaTX, inc. Cambridge (2023).
Summary
Keywords
mRNA therapeutics, mRNA vaccine, mRNA delivery, protein replacement therapy, mRNA design and synthesis, mRNA patents and clinical trials
Citation
Heendeniya SN, Chen S, Bhatti S, Zahra QUA, Rahimizadeh K, Poudel BH, Wilton SD and Veedu RN (2025) Beginning of a new era of synthetic messenger RNA therapeutics: Comprehensive insights on mRNA drug design, development and applications. Exp. Biol. Med. 250:10784. doi: 10.3389/ebm.2025.10784
Received
05 August 2025
Revised
16 October 2025
Accepted
21 October 2025
Published
12 December 2025
Volume
250 - 2025
Updates
Copyright
© 2025 Heendeniya, Chen, Bhatti, Zahra, Rahimizadeh, Poudel, Wilton and Veedu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rakesh N. Veedu, r.veedu@murdoch.edu.au
ORCID: Saadia Bhatti, orcid.org/0009-0000-7754-7221
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.