Abstract
Human respiratory syncytial virus (hRSV) is one of the most prevalent viruses infecting children globally. In this study, we employed the RT-RPA with CRISPR/Cas12a detection methodology to detect and differentiate RSV-A and RSV-B, particularly in resource-limited settings. The detection limit for RSV-A and RSV-B was approximately 102 and 103 copies/reaction, respectively. The assay revealed 100% specificity in detecting both RSV-A and RSV-B. Diagnostic accuracy was 90.32 and 93.55% for RSV-A and RSV-B, respectively, compared to RT-qPCR. These data indicate a proficient strategy for RSV screening, demonstrating promise for prospective applications in detecting diverse viral infections.
Impact statement
This study presents a novel RSV detection method using RT-RPA and CRISPR/Cas12a, achieving high sensitivity and specificity. With detection limits of 102 and 103 copies/reaction for RSV-A and RSV-B, respectively, and diagnostic accuracy comparable to RT-qPCR, it offers a reliable, resource-efficient alternative for accurate RSV screening.
Introduction
The human respiratory syncytial virus (hRSV) is an enveloped (−) ssRNA virus belonging to the Paramyxoviridae family and is classified in the genus Pneumovirus. RSV has a single serotype divided into two major subgroups, RSV-A and RSV-B, each comprising multiple genotypes [1–3]. The replication of hRSV occurs in both the upper and lower airways and is commonly disseminated through direct contact or aerosol inhalation [2, 4]. The predominant clinical presentation of RSV infection observed in children is bronchiolitis, a pathological condition affecting the lower respiratory tract, marked by airway obstruction. This condition may progress to complications such as pneumonia, respiratory failure, and mortality [5, 6].
Presently, preventive strategies rely on avoiding risk factors, patient isolation, and vaccination. However, existing vaccines primarily offer inadequate protection against RSV infection in older adults and high-risk individuals. Consequently, rapid diagnosis of RSV infection is essential to track and reduce the spread of diseases. The World Health Organization (WHO) advocates the utilization of a virus testing approach employing quantitative reverse transcription-polymerase chain reaction (qRT-PCR) conducted on throat or nasopharyngeal swabs (NP) [7, 8]. However, the use of qRT-PCR remains restricted because of the high cost of the devices, the need for a specialized molecular lab and specialists, and the 4-h detection turnaround time.
Interestingly, the U.S. Food and Drug Administration (FDA) recently approved the CRISPR-based diagnostic approach for more straightforward and rapid diagnosis [9]. This approach integrates the CRISPR/Cas system with recombinase polymerase amplification (RPA) to amplify and identify target regions within DNA sequences [10, 11]. The CRISPR-Cas12a technology, with its portable, rapid, and precise characteristics, holds immense promise in nucleic acid diagnostics, potentially transforming the field of virology and molecular diagnostics [12].
CRISPR-Cas12a, a CRISPR nuclease, possesses the capability to selectively degrade designated double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) that is complementary to the CRISPR RNA (crRNA). This process involves the utilization of the RuvC domain, resulting in the creation of staggered ends in the target DNA (cis-cleavage activity) after recognition of the protospacer adjacent motif (PAM) (5′-TTTV-3′) [13, 14]. After identifying the corresponding sequences to the CRISPR RNA (crRNA), the Cas protein cleaves the specific target. A matching DNA sequence triggers the collateral effect (trans-cleavage activity) of Cas12a. This activation separates the fluorescent reporter from the quencher, illuminating the fluorescent signal [15].
Despite extensive research on the CRISPR-Cas12a technology for the expeditious and precise detection of human respiratory viruses, there remains a constrained capacity for nucleic acid identification of RSV in clinical samples in relation to both subtypes, A and B. This limitation underscores the need for a more advanced and specific diagnostic tool. Therefore, the objective of this study is to establish an RT-RPA with a CRISPR/Cas12a-based technique specifically designed to identify RSV-A and RSV-B, a breakthrough that could revolutionize RSV diagnostics.
Materials and methods
Design and selection of primers and crRNA for specific RSV detection
Primers specific to the glycoprotein (G) genes of the human respiratory syncytial virus (RSV) and CRISPR RNA (crRNA) were meticulously designed to target the conserved region, and the sequences were selected from the NCBI database1. In designing crRNA, the selected sequences were adjacent to the PAM sequence (TTTV) [13], and the scaffold and T7 promoter sequence were then added to the 5′-end. This careful design process ensures the specificity and accuracy of our diagnostic tool. All primers and crRNAs used are presented in Table 1.
TABLE 1
| Assay | Primer name | Sequence (5´ ⟶ 3′) | Length (bp) | Product size (bp) |
|---|---|---|---|---|
| RT-RPA | RSVA-G-F282 | ATACCTCACCCAGAATCCCCAGCTTGGAAT | 30 | 391 |
| RSVA-G-R672 | GGTTTGAGGTTTGRGATCTTTTTTGGTTGTCTT | 33 | ||
| RSVB-G-F39 | CACTGCCAGTACTCTAGAAAAGACCTGGGA | 30 | 384 | |
| RSVB-G-R422 | CTGCCTTTGGTTTGTGCTGTTGTATGGTGT | 30 | ||
| crRNA | RSVA-G598 | UAAUUUCUACUAAGUGUAGAUCAGGUUUUUUGUUUGGUAUU | 41 | |
| RSVB-G154 | UAAUUUCUACUAAGUGUAGAUGCAAUGAUAAUCUCAACCUC | 41 |
Oligonucleotides used in the study.
Note: The underline represents the spacer sequence.
crRNA construction and purification
For crRNA construction, the annealing reaction consisted of 10 μL of a 2.5 μL of 10x ligation buffer, 10 μM of DNA template for crRNA, 10 μL of a 10 μM T7 promoter primer, and 2.5 μL of DEPC. The annealing process was as follows: 95°C for 3 min, 65°C for 3 min, 42°C for 5 min, and 37°C for 45 min. Subsequently, crRNA transcription was executed utilizing the RiboMAX™ Large Scale RNA Production System (Promega, United States), following the manufacturer’s prescribed protocol. Post-transcription, the crRNA, consisting of approximately 40 nucleotides, underwent purification employing a miRNA isolation kit (Geneaid, Taiwan) and was subsequently quantified using the Qubit™ microRNA Assay Kit (Thermo Fisher Scientific, United States).
RT-RPA reaction setup and protocol
A commercial reverse transcription recombinase polymerase amplification (RT-RPA) kit (TwistDx, UK) was employed to perform the isothermal amplification of the RSV-A and RSV-B genes. In brief, the RT-RPA reaction was constituted by combining 29.5 μL of rehydration buffer, 2.4 μL of a 10 μM forward primer, 2.4 μL of a 10 μM reverse primer, 1 μL of 200 U/μL Reverse Aid RT (Thermo Fisher Scientific, United States), 4.2 μL of DEPC, and 1 μL of the RNA template. Subsequently, 280 nM of MgOAC was introduced to the cap, and the mixture was vortexed and spun down for 2 min. The mixture was incubated in a mini heating dry bath (Major Science, United States) at 39°C for 30 min, followed by a subsequent heat inactivation step at 75°C for 5 min.
CRISPR/Cas12a cleavage assay procedure
Next, the CRISPR/Cas12a cleavage assay was conducted within a 15 μL reaction system. The mixture comprised 1.5 μL of 10x NEB buffer 2, 1.5 μL of 300 nM crRNA, 0.5 μL of 1 μM Cas12a (New England Biolabs, United States), 0.5 μL of 6 μM ssDNA probe labeled with FAM and BHQ1, 10 μL of DEPC, and 1 μL of RT-RPA template. Subsequently, the reaction mixtures underwent incubation in a mini heating dry bath (Major Science, United States) at 39°C for 15 min. The fluorescent signal was observed on a BluPAD Dual LED Blue/White Light Transilluminator (Bio-Helix, Taiwan) by the naked eye. The assay was conducted in triplicate, and the findings were based on consensus interpretations derived from three interpreters. The criteria for interpretation by the naked eye include the absence of fluorescence in negative samples, and a clear fluorescence or color change in positive samples. Positive results should be visibly distinguishable from negative controls, with the test sample showing a stronger signal than the background or controls.
Determining the limit of detection for RT-RPA and CRISPR-Cas12a
To determine the limit of detection, standard RNA was generated using RiboMAX™ Large Scale RNA Production System (Promega, United States) according to the manufacturer’s protocol. Then, DNase I (Thermo Fisher Scientific, United States) was used to eliminate the DNA templates. Next, the synthesized standard RNA was purified and quantified using a miRNA isolation kit (Geneaid, Taiwan) and Qubit™ microRNA Assay Kit (Thermo Fisher Scientific, United States), respectively. Using a 10-fold serial dilution, synthetic standard RNAs with copy numbers ranging from 107 to 10 copies/μL were prepared. The limit of detection was determined by the RT-RPA followed by CRISPR-Cas12a detection. The fluorescent signal was observed on a BluPAD Dual LED Blue/White Light Transilluminator (Bio-Helix, Taiwan) by the naked eye.
Cross-reactivity testing with various respiratory viruses
Next, cross-reactivity testing was conducted on diverse human respiratory viral samples, including influenza viruses A (A/H1 and A/H3), influenza viruses B (B/Yam and B/Vic), human bocavirus (HBoV), coronavirus (CoV), human metapneumovirus (HMPV), human parainfluenza virus (HPIV), and human rhinovirus (HRV). This assessment was carried out utilizing RT-RPA and CRISPR-Cas12a detection methodologies (n = 10 for each).
Calculation of sensitivity, specificity, and accuracy
The sensitivity, specificity, and accuracy were calculated using the web-based diagnostic test evaluation calculator MedCalc software2 [16]. This web-based tool was utilized to calculate these metrics, ensuring a comprehensive analysis of the test performance. The formulas for calculating sensitivity, specificity, and accuracy are as follows:
RNA sample collection and verification
The RNA samples were obtained from the Center of Excellence in Clinical Virology, Chulalongkorn University. Briefly, nasopharyngeal swab samples (n = 93) were collected from patients with influenza-like illness (ILI). The viral RNA was extracted using a GenUP™ Virus RNA kit (Biotech Rabbit, German). The RSV-positive sample was verified by RT-qPCR as described in the previous study [17] (cut-off at Ct ≤ 38).
Results and discussion
A recently developed approach for pathogen detection incorporating isothermal amplification techniques (such as RPA) coupled with fluorescent detection utilizing the CRISPR-Cas system [10, 18, 19] has been developed and shown to be useful for higher-sensitivity nucleic acid pathogen identification [11, 20]. Our experimental protocol procedure took approximately 90 min, covering the entire process from RNA extraction to fluorescent detection (Figure 1). Meanwhile, the qRT-PCR revealed an approximately 4-h turn-around time and required a specialized molecular laboratory, expensive equipment, and well-trained staff.
FIGURE 1
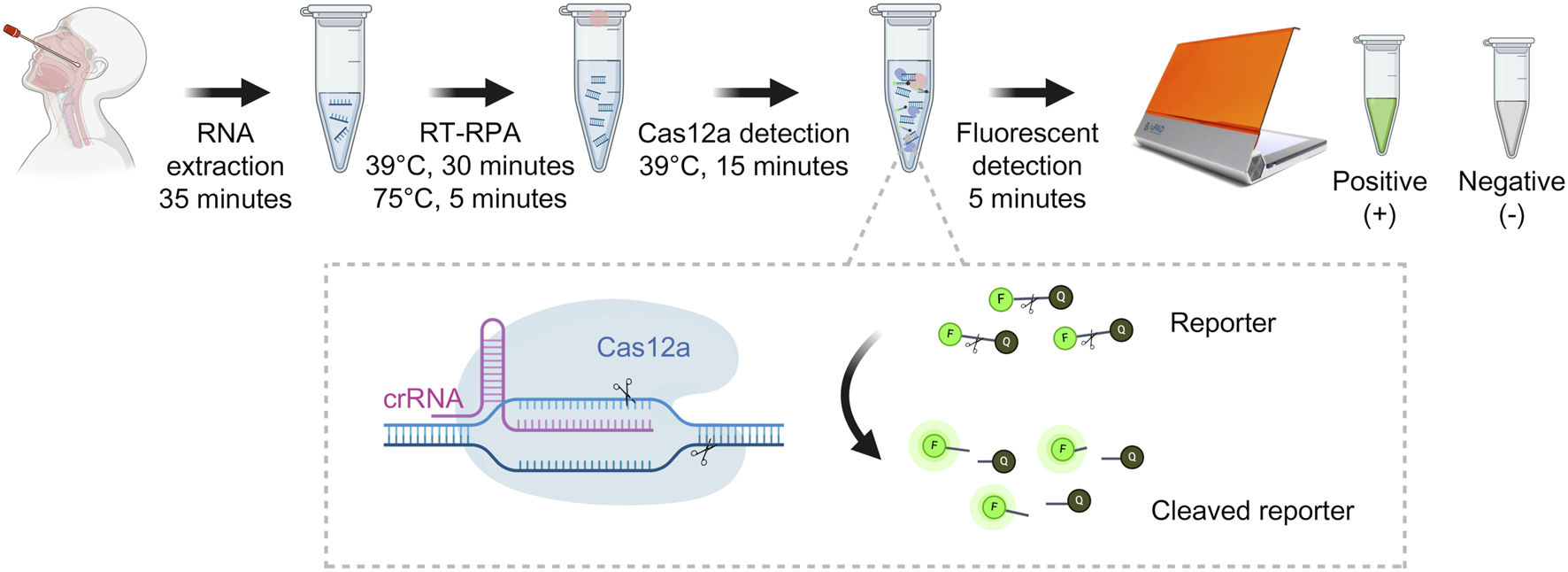
Detection of RSV-A and RSV-B based on RT-RPA and CRISPR-Cas12a (This figure was created with Biorender.com).
A previous study has shown that RSV can be detected using the RPA coupled with the CRISPR-Cas approach at 104 copies/mL [21]. As illustrated in the results (Figure 2A), the LOD for RSV-A and RSV-B were estimated to be around 102 and 103 copies per reaction, respectively. The outcomes demonstrated that the primers and crRNA used in this investigation exhibited notable specificity against RSV-A and RSV-B viruses (Figure 2B). No cross-reactivity was discerned with other respiratory viruses (Figure 2B). Interestingly, our study can discriminate between the RSV-A and RSV-B subtypes (Figure 2C). In addition, RT-RPA with CRISPR-Cas12a assay was performed using RSV-A primers on RSV-B targets, and vice versa. The results showed that the primers could not amplify across subtypes (Supplementary Figure S1). The representative detection results were shown in Figure 2D.
FIGURE 2
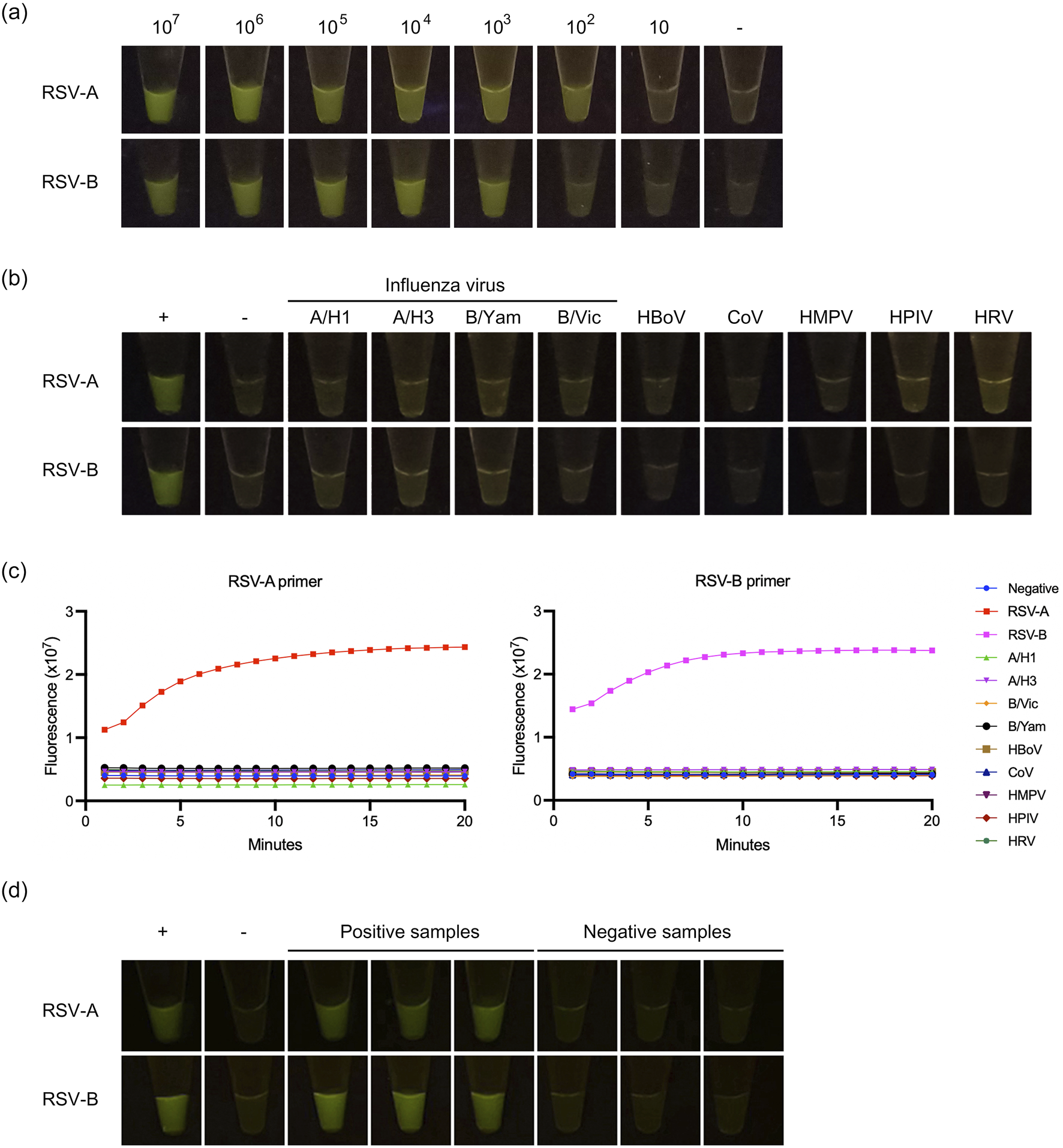
Detection of RSV-A and RSV-B based on RT-RPA and CRISPR-Cas12a. (A) The limit of detection. (B) The cross-reactivity testing against influenza viruses A (A/H1 and A/H3), influenza viruses B (B/Yam and B/Vic), human bocavirus (HBoV), coronavirus (CoV), human metapneumovirus (HMPV), human parainfluenza virus (HPIV), and human rhinovirus (HRV). (C) Cross-reactivity testing against other respiratory viruses was performed by qPCR using RT-RPA + CRISPR with RSV-A primers and RT-RPA + CRISPR with RSV-B primers. Dot plots show fluorescence intensity measurements taken from 1 to 15 min. (D) The representative of positive and negative detection.
In this study, the RSV-positive sample was verified by RT-qPCR. The results showed 39 and 27 RSV-A and RSV-B positive samples, respectively. Prior investigations indicated that RSV-A exhibited specificity and sensitivity values of 73.08% and 90%, respectively, while RSV-B demonstrated specificity and sensitivity figures of 42.86% and 93.33%, respectively [22]. Our study suggests a modest enhancement in the specificity (100%) of RSV-A and RSV-B detection compared to the gold-standard detection outcomes from RT-qPCR, as shown in Table 2. However, the current study demonstrates lower sensitivity, with values of 76.92% and 77.78% for RSV-A RSV-B, respectively (Table 2). In the case of RSV-A, the findings revealed 30 true positives, 0 false positives, 9 false negatives, and 54 true negatives. Conversely, for RSV-B, the results indicated 21 true positives, 0 false positives, 6 false negatives, and 66 true negatives. The undetected positive samples were associated with high Ct (cycle threshold) values, which could indicate a lower viral load in these samples. Furthermore, since multiple freeze-thaw cycles and storage may have caused RNA degradation. Additionally, the diagnostic accuracy for RSV-A and RSV-B was reported as 90.32% and 93.55%, respectively.
TABLE 2
| Parameters | RSV-A | RSV-B |
|---|---|---|
| Total samples | 93 | 93 |
| True positive | 30 | 21 |
| True negative | 54 | 66 |
| False positive | 0 | 0 |
| False negative | 9 | 6 |
| Sensitivity | 76.92% | 77.78% |
| Specificity | 100% | 100% |
| Diagnostic accuracy | 90.32% | 93.55% |
The performance of RT-RPA and CRISPR-Cas12a based assay.
However, this research still has some limitations, including the challenges related to nonspecific amplification, variable sensitivity, isothermal conditions, guide RNA specificity, and the subjective nature of the readout. These highlight the importance of ongoing research and optimization for their practical and reliable application in diagnostics.
In conclusion, the benefits of this study include simplicity, quick turnaround time, affordability, and applicability in resource-constrained places. The assays would be helpful and appealing for systematic retesting of individuals and large-scale screening across many communities to isolate patients and prevent the spread of RSV.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The experimental procedures in this investigation were approved by the Institutional Review Board, Faculty of Medicine, Chulalongkorn University (IRB No. 0352/67).
Author contributions
AK: Formal Analysis, Methodology, Visualization, Writing – original draft. JN: Methodology, Writing – review and editing. NC: Methodology, Writing – review and editing. PN: Methodology, Writing – review and editing. OM: Methodology, Writing – review and editing. PK: Supervision, Writing – review and editing. SiP: Methodology, Writing – review and editing. YP: Supervision, Writing – review and editing. SuP: Conceptualization, Funding acquisition, Supervision, Writing – review and editing. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Research Grants for Talented Mid-Career Researchers, The National Research Council of Thailand (NRCT) (N41A640077); the Thailand Science Research and Innovation Fund, Chulalongkorn University (HEAF67300057); Ratchadapisek Somphot Fund, Faculty of Medicine, Chulalongkorn University (MDCU RA-MF-21/67); and Ratchadapisek Somphot Fund for Postdoctoral Fellowship, Chulalongkorn University.
Acknowledgments
We would like to thank the Center of Excellence in Clinical Virology, Faculty of Medicine, Chulalongkorn University, for kindly providing the samples.
Conflict of interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.ebm-journal.org/articles/10.3389/ebm.2025.10387/full#supplementary-material
References
1.
MufsonMAOrvellCRafnarBNorrbyE. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol (1985) 66(Pt 10):2111–24. 10.1099/0022-1317-66-10-2111
2.
CollinsPLFearnsRGrahamBS. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol (2013) 372:3–38. 10.1007/978-3-642-38919-1_1
3.
JorqueraPAAndersonLTrippRA. Human respiratory syncytial virus: an introduction. Methods Mol Biol (2016) 1442:1–12. 10.1007/978-1-4939-3687-8_1
4.
KrauseCI. The ABCs of RSV. The Nurse Pract (2018) 43:20–6. 10.1097/01.npr.0000544277.74514.55
5.
BarrRGreenCASandeCJDrysdaleSB. Respiratory syncytial virus: diagnosis, prevention and management. Ther Adv Infect Dis (2019) 6:2049936119865798. 10.1177/2049936119865798
6.
PiedimonteGPerezMK. Respiratory syncytial virus infection and bronchiolitis. Pediatr In Rev (2014) 35:519–30. 10.1542/pir.35.12.519
7.
TeirlinckACBrobergEKStuwitz BergACampbellHReevesRMCarnahanAet alRecommendations for respiratory syncytial virus surveillance at the national level. Eur Respir J (2021) 58:2003766. 10.1183/13993003.03766-2020
8.
ToddAKCostaAMWallerGDaleyAJBarrIGDengYM. Rapid detection of human respiratory syncytial virus A and B by duplex real-time RT-PCR. J Virol Methods (2021) 294:114171. 10.1016/j.jviromet.2021.114171
9.
Puig-SerraPCasado-RosasMCMartinez-LageMOlalla-SastreBAlonso-YanezATorres-RuizRet alCRISPR approaches for the diagnosis of human diseases. Int J Mol Sci (2022) 23:1757. 10.3390/ijms23031757
10.
MayuramartONimsamerPRattanaburiSChantaravisootNKhongnomnanKChansaenrojJet alDetection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Exp Biol Med (Maywood) (2021) 246:400–5. 10.1177/1535370220963793
11.
NimsamerPMayuramartORattanaburiSChantaravisootNSaengchoowongSPuenpaJet alComparative performance of CRISPR-Cas12a assays for SARS-CoV-2 detection tested with RNA extracted from clinical specimens. J Virol Methods (2021) 290:114092. 10.1016/j.jviromet.2021.114092
12.
WangBWangRWangDWuJLiJWangJet alCas12aVDet: a CRISPR/cas12a-based platform for rapid and visual nucleic acid detection. Anal Chem (2019) 91:12156–61. 10.1021/acs.analchem.9b01526
13.
PaulBMontoyaG. CRISPR-Cas12a: functional overview and applications. Biomed J (2020) 43:8–17. 10.1016/j.bj.2019.10.005
14.
DroninaJSamukaite-BubnieneURamanaviciusA. Towards application of CRISPR-Cas12a in the design of modern viral DNA detection tools (Review). J Nanobiotechnology (2022) 20:41. 10.1186/s12951-022-01246-7
15.
WangYYangTLiuGXieLGuoJXiongW. Application of CRISPR/Cas12a in the rapid detection of pathogens. Clinica Chim Acta (2023) 548:117520. 10.1016/j.cca.2023.117520
16.
SchoonjansFZalataADepuydtCEComhaireFH. MedCalc: a new computer program for medical statistics. Computer Methods Programs Biomed (1995) 48:257–62. 10.1016/0169-2607(95)01703-8
17.
ThongpanISuntronwongNVichaiwattanaPWanlapakornNVongpunsawadSPoovorawanY. Respiratory syncytial virus, human metapneumovirus, and influenza virus infection in Bangkok, 2016-2017. PeerJ (2019) 7:e6748. 10.7717/peerj.6748
18.
MaoXXuMLuoSYangYZhongJZhouJet alAdvancements in the synergy of isothermal amplification and CRISPR-cas technologies for pathogen detection. Front Bioeng Biotechnol (2023) 11:1273988. 10.3389/fbioe.2023.1273988
19.
LiXZhuSZhangXRenYHeJZhouJet alAdvances in the application of recombinase-aided amplification combined with CRISPR-Cas technology in quick detection of pathogenic microbes. Front Bioeng Biotechnol (2023) 11:1215466. 10.3389/fbioe.2023.1215466
20.
ChenJSMaEHarringtonLBDa CostaMTianXPalefskyJMet alCRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science (2018) 360:436–9. 10.1126/science.aar6245
21.
HongdanGYaoDQiangCMengHXiaorongLZhihaoXet alA multiplex recombinase polymerase amplification assay combined with CRISPR/Cas12a for the detection of respiratory syncytial virus and respiratory adenovirus. J Int Med Res (2024) 52:3000605231223083. 10.1177/03000605231223083
22.
GongLWangXLiZHuangGZhangWNieJet alIntegrated trinity test with RPA-CRISPR/cas12a-fluorescence for real-time detection of respiratory syncytial virus A or B. Front Microbiol (2022) 13:819931. 10.3389/fmicb.2022.819931
Summary
Keywords
CRISPR-Cas12a, detection, isothermal amplification, RT-RPA, respiratory syncytial virus
Citation
Khamwut A, Nimnual J, Chomta N, Nimsamer P, Mayuramart O, Kaewsapsak P, Pasittungkul S, Poovorawan Y and Payungporn S (2025) Detection of respiratory syncytial virus based on RT-RPA and CRISPR-Cas12a. Exp. Biol. Med. 250:10387. doi: 10.3389/ebm.2025.10387
Received
25 September 2024
Accepted
22 April 2025
Published
01 May 2025
Volume
250 - 2025
Updates
Copyright
© 2025 Khamwut, Nimnual, Chomta, Nimsamer, Mayuramart, Kaewsapsak, Pasittungkul, Poovorawan and Payungporn.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunchai Payungporn, sp.medbiochemcu@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.